- Journal List
- HHS Author Manuscripts
- PMC6689410

Selenium Deficiency Is Associated with Pro-longevity Mechanisms
Sun Hee Yim
1Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA 02115, USA
2Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA
Clary B. Clish
2Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA
Vadim N. Gladyshev
1Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA 02115, USA
2Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA
3Lead Contact
AUTHOR CONTRIBUTIONS
Conceptualization, S.H.Y. and V.N.G.; Methodology, S.H.Y. and C.B.C.; Investigation, S.H.Y., C.B.C., and V.N.G.; Resources, S.H.Y., C.B.C., and V.N.G.; Writing – Original Draft, S.H.Y. and V.N.G.; Writing – Review & Editing, S.H.Y., V.N.G., and C.B.C.Associated Data
- Data Availability Statement
All primary LC-MS metabolite profiling data, RNaseq result, miRNA data are available as supplemental files. Raw RNA-seq data reported in this paper were submitted to the Gene Expression Omnibus database available at https://www.ncbi.nlm.nih.gov/geo/ under the accession code {"type":"entrez-geo","attrs":{"text":"GSE129138","term_id":"129138","extlink":"1"}}GSE129138.
SUMMARY
Selenium (Se) is an essential trace element because of its presence in selenoproteins in the form of selenocysteine residue. Both Se deficiency, which compromises selenoprotein functions, and excess Se, which is toxic, have been associated with altered redox homeostasis and adverse health conditions. Surprisingly, we found that, although Se deficiency led to a drastic decline in selenoprotein expression, mice subjected to this dietary regimen for their entire life had normal lifespans. To understand the molecular mechanisms involved, we performed systemic analyses at the level of metabolome, transcriptome, and microRNA profiling. These analyses revealed that Se deficiency reduced amino acid levels, elevated mononucleotides, altered metabolism, and activated signaling pathways linked to longevity-related nutrient sensing. The data show that the metabolic control associated with nutrient sensing coordinately responds to suppressed selenoprotein functions, resulting in normal lifespan under Se deficiency.
In Brief
Yim et al. report that selenium deficiency in mice is associated with pro-longevity mechanisms because of reduced amino acid levels and altered nutrient signaling.
INTRODUCTION
Selenium (Se) is an essential trace element in mammals because of its occurrence in Se-containing proteins (selenoproteins) in the form of selenocysteine (Sec) encoded by UGA codons. There are 25 selenoprotein genes in humans and 24 in mice (Kryukov et al., 2003). Several of these genes (Txnrd1, Txnrd2, and GPx4) and the gene for the tRNA for Sec are essential for embryonic development. Selenoproteins have important roles in redox homeostasis, and the mammalian thioredoxin (Trx) system is fully dependent on Se because both thioredoxin reductases (cytosolic Txnrd1 and mitochondrial Txnrd2) are selenoproteins (Jakupoglu et al., 2005; Matsui et al., 1996; Nonn et al., 2003; Soerensen et al., 2008). When Se supplies are low, selenoprotein levels are decreased in an organ- and selenoprotein-specific manner: although Se deficiency may lead to a loss of more than 90% Se in liver and kidney, brain and testes are resistant to that dietary regimen, wherein Se is redistributed within the body toward those organs (Hill et al., 2003; Nakayama et al., 2007).
Both Se excess (Institute of Medicine, 2000; MacFarquhar et al., 2010) and deficiency (Rayman, 2012) are associated with health risk in humans and mice. The association between Se status and human disease is evidenced by clinical trials and molecular analyses (Christen et al., 2015; Vinceti et al., 2014), but a clear picture has yet to emerge because changes in the levels of certain selenoproteins may both promote disease and protect against it (Hatfield et al., 2009; Lu and Holmgren, 2009; Rayman, 2012). The best-studied human disease affected by Se availability is cancer (Allen et al., 2008; Dennert et al., 2011; Erdman et al., 2012; Klein et al., 2011; Kristal et al., 2014; Lippman et al., 2009; Rayman, 2012; Reid et al., 2008), but the status of this element has also been associated with diabetes and other metabolic disorders (Hellwege et al., 2014; Ogawa-Wong et al., 2016; Rayman and Stranges, 2013; Seale et al., 2015), as well as with other adverse conditions and oxidative stress (Takata et al., 2012).
Dietary selenite is directly reduced to selenide, whereas selenomethionine can be either metabolized through the transsulfuration pathway or used in place of methionine for protein synthesis, methylation, and other processes dependent on this amino acid (Anan and Ogra, 2013). Interestingly, in addition to their primary Trx substrates, thioredoxin reductases (Txnrds) display broad substrate specificities and reduce various low-molecular-weight compounds, including Se compounds. This indicates that the functions of these enzymes may interact with those of low molecular weight metabolites in redox homeostasis and Se metabolism (Arnér, 2009; Eriksson et al., 2015; Prigge et al., 2017; Scranton and McDermott, 1997). The optimum dietary Se concentration varies according to environment, to the forms of Se intake, to genotype, and to age (Hurst et al., 2010). The estimated Se requirement in mice and rats is 150 μg/kg diet (0.15 ppm Se) in the form of selenite. The current recommended dietary allowance (RDA) for humans is 55 μg (0.7 μmol)/day, and selenosis, a condition associated with Se toxicity, is observed with greater than 400 μg (5.1 μmol)/day, which is considered a tolerable upper intake level (Institute of Medicine, 2000). Based on these considerations, several Se dietary regimens have been devised, and we employed them in the current study. These include diets (1) deficient in dietary Se (based on Se-deficient torula yeast diet), i.e., 0 ppm Se diet; (2) the 0.1 ppm Se diet, in which the level of Se corresponds to the minimal amount needed to maximize selenoprotein expression, i.e., further increases in dietary Se do not support increases in selenoprotein levels; (3) the 0.4 ppm Se diet, which corresponds to the diet enriched in Se, e.g., through supplementation in mice or a pill in humans; and (4) the 2.25 ppm Se diet, which corresponds to a high Se level, representing a lower bound of Se toxicity.
Mammalian aging is accompanied by diverse metabolic changes. One of the most successful and well-understood interventions against the onset of aging is limitation in nutrient intake, e.g., life-long caloric restriction can increase the lifespan of rodents by up to 50% (de Cabo et al., 2014). Key nutrient-sensing pathways associated with extended longevity involve mechanistic target of rapamycin (mTOR), sirtuins, and AMP kinase (AMPK). Among these, mTOR senses cellular amino acid levels and activates cell growth in response to nutrients (e.g., amino acids), growth factors (e.g., insulin and insulin-like growth factor) and cellular energy status (ATP). The levels and ATP:AMP ratio are critical for energy homeostasis, with AMPK being a fundamental regulator of energy sensing (Hardie, 2011).
There have been few studies on the association between dietary Se levels and mammalian lifespan. However, the links between Se (or other trace elements) and this trait are not well understood. A recent epidemiology analysis of the effect of long-term Se supplementation on mortality pointed to a possibility that the lower dose of Se may be associated with decreased mortality, although the effect disappeared after treatment cessation (Rayman, 2017). In rodents, Selenoprotein P (Selenop)-deficient mice showed elevated endurance after exercise training, enhanced reactive oxygen species (ROS) production, and increased AMPK phosphorylation (Misu et al., 2017). Increased lifespan and reduced age-associated pathology were observed in response to a life-long reduction in GPx4 levels (using GPx4 heterozygous knockout mice) because of alterations in sensitivity of tissues to apoptosis (Ran et al., 2007). Interestingly, short-lived telomerase RNA knockout (G3 Terc−/−) mice subjected to Se deficiency showed poor health, but that treatment promoted longevity (Wu et al., 2017). In addition to the regulation of selenoproteins and their transcript levels (Raines and Sunde, 2011), Se deficiency may directly and indirectly affect other cellular components, e.g., the levels of certain metabolites change in response to Se deficiency or toxicity in humans, mice, and yeast (Arnaudguilhem et al., 2012; Mickiewicz et al., 2014; Sinha et al., 2016). In this regard, a more systemic approach is needed to obtain insights into regulation of cellular and organismal processes involving dietary Se and its potential links with nutrient sensing. Here, we directly examined how dietary Se regulates metabolism at the molecular level and how it affects mouse lifespan.
RESULTS
Dietary Se Regulates Selenoprotein Expression but Does Not Affect Mouse Lifespan
We subjected 8-week old, C57BL/6J, male mice to diets supplemented with 0, 0.1, 0.4, and 2.25 ppm Se in the form of sodium selenite. Hereafter, the 0 ppm Se group has been designated as Se-deficient, the 0.1- and 0.4 ppm Se groups as Se-sufficient, and the 2.25 ppm Se as toxic. Because of active processes of Se retention in response to Se deficiency, 4 weeks of treatment is typically needed to adjust and stabilize the expression of selenoproteins and other cellular components (Barnes et al., 2009; Bodnár et al., 2016; Hill et al., 2012; Hrdina et al., 2009; Olson et al., 2010; Sunde et al., 2009; Wu et al., 2016). Therefore, the mice were subjected to all diets for 5 weeks. We first evaluated how the four Se diets influence selenoprotein expression (Figure 1A). The levels of methionine sulfoxide reductase B1 (MsrB1) and glutathione peroxidase 1 (GPx1) were dramatically reduced in liver by the 0 ppm Se diet. Txnrd1 (TR1) expression was moderately reduced by Se deficiency, whereas Txnrd2 (TR3) and its non-selenoprotein substrate Trx2 were not affected. In contrast to the liver, the levels of MsrB1 (a stress-related selenoprotein) and Txnrd2 (a housekeeping selenoprotein) in the brain were not affected by Se deficiency. These data showed that Se deficiency robustly affected selenoprotein expression, consistent with previous data on regulation of selenoprotein function by dietary Se (Novoselov et al., 2010).
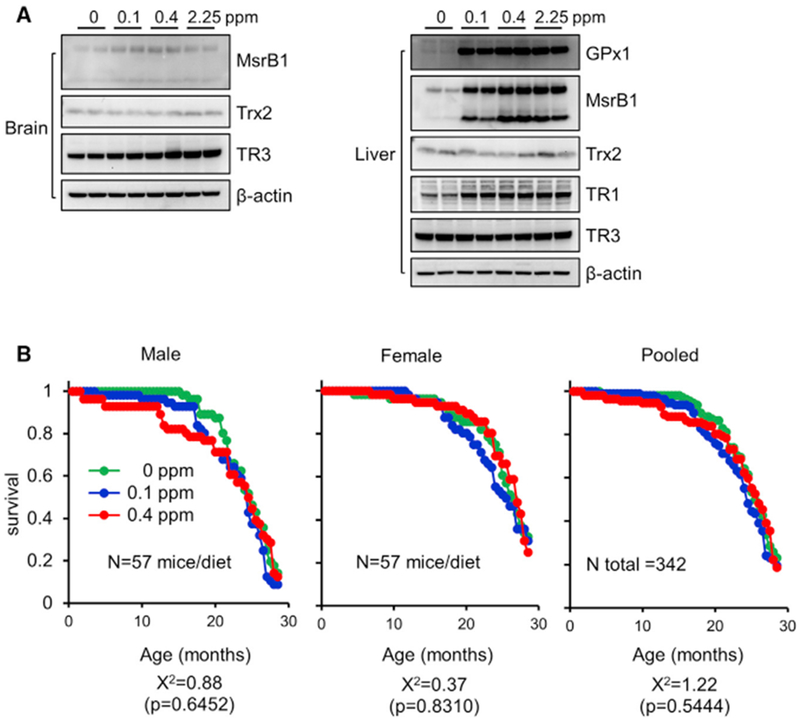
(A) Western blot analyses of brain (left) and liver (right) of mice subjected to various Se diets (0, 0.1, 0.4, and 2.25 ppm) for 5 weeks (shown for two animals per group). Proteins probed are indicated on the right. Expression of MsrB1 (occurs in two forms represented by two bands), GPx1, and Txnrd1 (TR1) was reduced in liver by Se deficiency. Expression of Trx2 (not a selenoprotein) was analyzed as a control. β-actin served as a loading control.
(B) Kaplan-Meier survival curves for animals fed with diets containing 0,0.1, and 0.4 ppm Se. A total of 342 mice (57 animals/dietary group · sex) were subjected to designated Se diets for 28.5 months. The data are plotted for males (left), females (middle), and both sexes combined (pooled) (right). p values and chi-square distributions were derived from log-rank calculations. The chi-square distributions and p values are χ2 = 1.22 and p = 0.5444 when both male and female animals were combined.
To assess long-term consequences of altered selenoprotein expression, we examined the effect of Se diets on mouse lifespan. We fed 342 animals (equal numbers of males and females) with 0, 0.1, or 0.4 ppm Se diets and followed their lifespan (Figure 1B). Kaplan-Meier survival analysis and log-rank test for the equality of survival distribution were used to assess the effect of Se; the χ2 values were 0.88 (p = 0.6452) and 0.37 (p = 0.8310) for males and females, respectively. For both males and females combined, the χ2 value was 1.22 (p = 0.5444), reflecting the lack of the effect of Se diets on lifespan. Overall, the data clearly showed that Se deficiency, despite the dramatic influence on selenoprotein levels, did not shorten the lifespan of mice. In fact, during the course of the study, mortality of mice on the Se-deficient diet was slightly less than it was for those on the 0.1 or 0.4 ppm diets, although the difference did not reach the level of statistical significance.
Dietary Se Regulates Liver and Brain Metabolomes
To understand how mice could have normal lifespan despite severe selenoprotein deficiency, we analyzed them by metabolite profiling (Data S1). Liver and brain of mice fed 0, 0.1, 0.4, and 2.25 ppm Se diets were profiled by both targeted and non-targeted methods to detect four groups of metabolites: (1) amines and cationic metabolites; (2) sugars, organic acids, purines, pyrimidines, and anionic metabolites; (3) lipids; and (4) free fatty acids, bile acids, lipid mediators, and metabolites of intermediate polarity. Using these methods, we measured 516 compounds of known identity (181 polar metabolites and 335 lipids and lipid mediators) and 10,000 and 15,000 features of unknown identity by polar metabolite and lipid profiling methods, respectively.
We then examined global changes in metabolite patterns in response to Se diets. Two interesting patterns emerged: (1) the metabolome was altered by Se deficiency more than by other diets, and (2) the liver responded much stronger to Se diets than the brain did. The 2.25 ppm Se diet, although having large excess of Se, showed few changes in metabolite levels in either the liver or the brain compared with other diets. This finding, when examined against regulation of selenoprotein levels (Figure 1A), shows that the metabolome principally changes in response to selenoprotein expression and not Se levels per se.
Se deficiency altered 10%–30% of amines and cationic metabolites in the liver and 7%–16% in the brain (0 ppm versus Se-sufficient groups), whereas Se toxicity affected 11% metabolites in the liver and 7% in the brain (0.4 versus 2.25 ppm comparison) (Figures 2A and S1A). In addition, 8%–38% (liver) and 6%–21% (brain) of sugars, organic acids, purines, and pyrimidines were changed by dietary Se. As above, fewer changes were observed in the comparison between 0.4- and 2.25 ppm diets (Figures 2B and S1B). Among the four metabolite profiling platforms, the fewest changes from dietary Se were observed in the lipid metabolome, i.e., 6%–22% (liver) and 3%–19% (brain) as being affected by dietary Se. Once again, the 0.4 versus 2.25 ppm comparison revealed few changes (6% in liver and 4% in brain) (Figures 2C and S1C). Among free fatty acids, bile acids, and lipid mediators, 11%–32% of metabolites changed in response to Se deficiency in the liver and 8%–31% in the brain (Figures 2D and S1D).
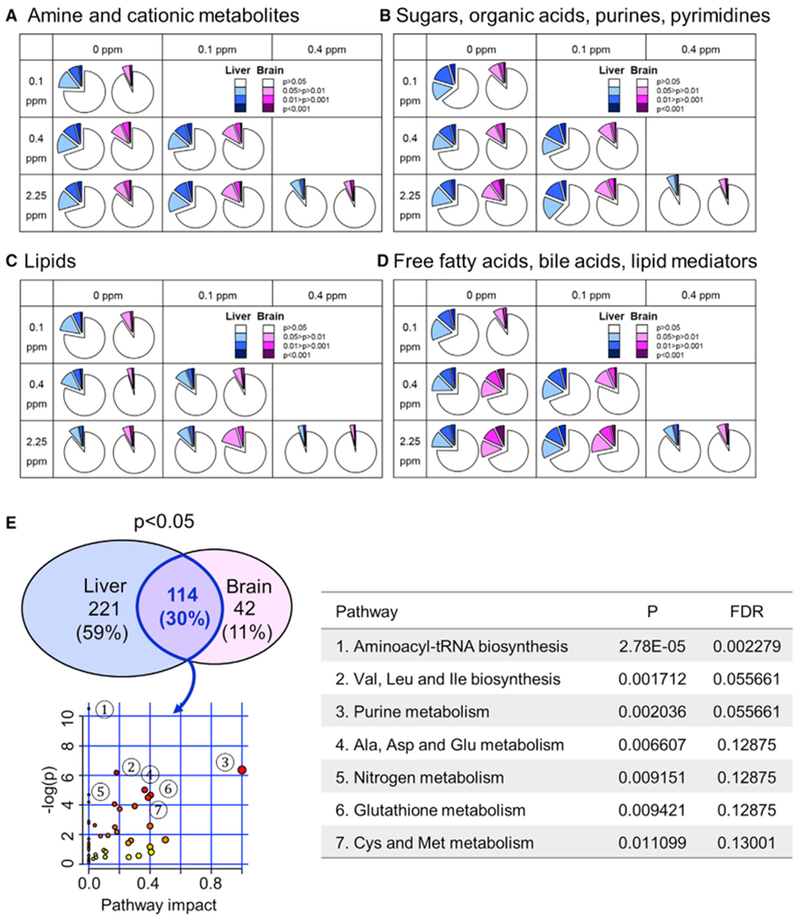
(A–D) Each panel represents a particular metabolite profiling platform, including (A) amine and cationic metabolites, (B) sugars, organic acids, purines, and pyrimidines, (C) lipids, and (D) free fatty acids, bile acids, and lipid mediators. and shows pairwise comparison between dietary groups using Student’s t test. Liver data are colored in blue and brain in pink. In each colored and expanded slice in the pie chart, darker color corresponds to higher statistical significance; p > 0.05 (i.e., no statistical difference) are shown in white. Highlighted areas in each slice in the pie chart indicate the fraction of metabolites affected by dietary Se in the comparison between indicated dietary groups.
(E) Metabolites commonly altered in liver and brain by dietary Se and the associated metabolic pathways. Three hundred thirty-five targeted metabolites in liver and 156 metabolites in brain that were altered by dietary Se at p < 0.05. Metabolites that changed in both liver and brain in response to any Se diet included 114 metabolites at p < 0.05 and 22 metabolites at p < 0.01. These 114 metabolites were further analyzed for pathway enrichment using MetaboAnalyst 4.0. The metabolome view contains all the matched pathways arranged by p values from the pathway enrichment analysis on the y axis as “−log(p).” The pathway impact values on the x axis were calculated from pathway topology analyses, which use the structure of a given pathway to evaluate the relative importance of genes and compounds based on their relative locations and based on the genes and metabolites that are mapped to KEGG metabolic pathways (Xia and Wishart, 2010). Significantly enriched molecular pathways are marked (Table S1).
Targeted metabolite profiling revealed 25 metabolites in the liver and one in the brain (p < 0.001) affected by Se deficiency and one metabolite in the liver and two in the brain (p < 0.001) affected by Se toxicity. Among 516 targeted metabolites, 186 metabolites (36%) were changed by Se deficiency and 62 (12%) by toxicity compared with Se-adequate diets in the liver (p < 0.05) (Figure S1E). Overall, dietary Se affected both liver and brain metabolomes, with the most changes elicited by Se deficiency.
Dietary Se Affects Redox Homeostasis via Methionine Metabolism
To better understand the effect of Se on the metabolome and the associated metabolic pathways, we focused on targeted (known) metabolites. 335 of them were altered (p < 0.05) in at least one comparison in the liver and 156 in the brain, including 114 of the 516 (22%) commonly regulated in both organs (p < 0.05) (Figure 2E). We examined the pathways involved with MetaboAnalyst 4.0 (Chong et al., 2018), determining whether common metabolites were affected in both organs in any of the six comparisons (0 versus 0.1 ppm, 0 versus 0.4 ppm, 0 versus 2.25 ppm, 0.1 versus 0.4 ppm, 0.1 versus 2.25 ppm, and 0.4 versus 2.25 ppm). After false discovery rate (FDR) adjustment, the pathways of amino acid biosynthesis and metabolism, nitrogen metabolism, and glutathione metabolism were the most enriched. Twenty-two metabolites (p < 0.01) that are commonly changed by Se in the liver and brain were associated with cysteine and methionine metabolism, ascorbate and aldarate metabolism, and amino acid biosynthesis (Table S1).
Among the metabolites affected by Se, several were associated with methionine metabolism and redox homeostasis, including methionine (Human Metabolome Database [HMDB]: 00696), methionine sulfoxide (HMDB: 02005), S-adenosyl methionine (SAM; HMDB: 01185), and 5-adenosylhomocysteine (SAH; HMDB: 00939) (Figure 3A). Methionine in the liver was decreased the most by Se deficiency. We also observed organ-specific changes in methionine sulfoxide, SAM, and SAH. For example, SAM levels in liver were increased 3-fold by Se deficiency, whereas its levels in the brain were not affected. These metabolites are associated with methionine metabolism that provides methyl groups for processes such as DNA methylation, RNA processing, modification of heavy metals, regulation of gene expression, and other metabolic pathways. These metabolites are also upstream of the transsulfuration pathway with its key sulfur-based redox metabolites, such as glutathione and cysteine (Figure 3B). Taken together, Se exerts its effects, including regulation of redox homeostasis and methionine metabolism, in an organ-specific manner.
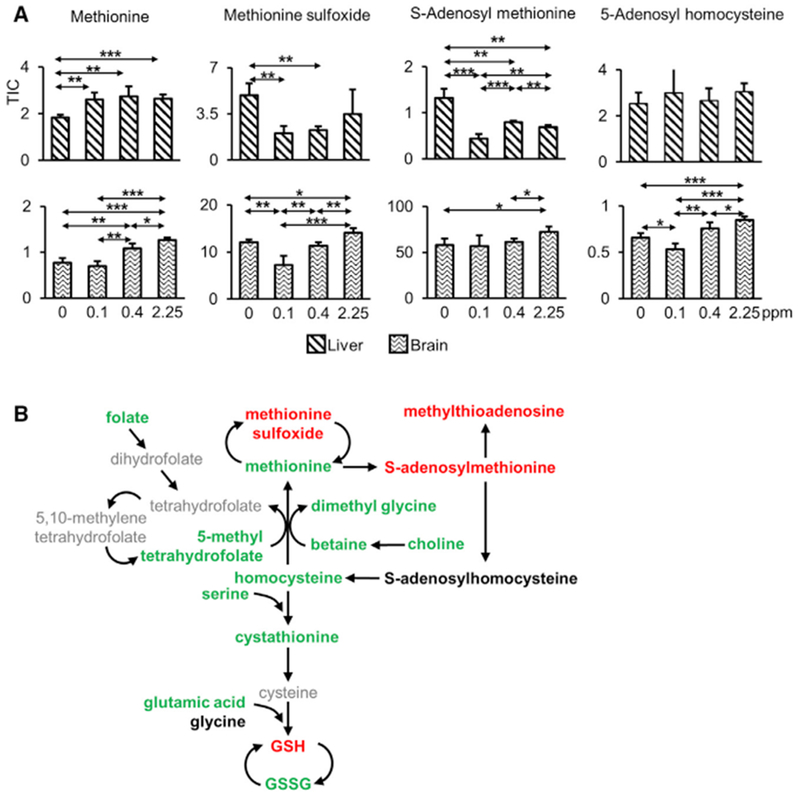
(A) Relative levels of four indicated metabolites are shown for liver (upper panels) and brain (lower panels). *p < 0.05, **p < 0.01, and ***p < 0.001 (two-tailed Student’s t test).
(B) Summary illustration of methionine metabolism regulated by dietary Se in liver. Targeted metabolites that were measured (in bold) and changed between Se deficiency and Se sufficiency are shown in color. Metabolites decreased by Se deficiency are shown in green, those that increased in red, and metabolites that are not altered are in black. Se deficiency decreases the levels of methyl donors, methionine, and its downstream metabolites, whereas it increased methionine sulfoxide. The ratio of glutathione (reduced) (GSH) to glutathione (oxidized) (GSSG) was increased under conditions of Se deficiency. The associated amino acids, methionine, serine, and glutamic acid were significantly decreased by Se deficiency. TIC represents total ion current.
Organ-Specific Changes in Nucleotide and Amino Acid Metabolism
Purines and pyrimidines serve as precursors of nucleic acids, energy carriers, components of co-enzymes, and cellular signaling molecules. The levels of mononucleotides, inosine 5’-monophosphate (IMP; HMDB: 00175), AMP (HMDB: 00045), guanosine 5’-monophosphate (GMP; HMDB: 01397), and uridine 5’-monophosphate (UMP; HMDB: 00288) were all elevated by Se deficiency (Figure 4A). Dietary Se also appeared to have a role in nucleotide catabolism. Pyrimidine catabolism showed organ-specific regulation by dietary Se (Figure 4B). Cytosine (HMDB: 00630), uracil (HMDB: 00300), and β-alanine (HMDB: 00056) were decreased by Se deficiency in liver, whereas N-carbamoyl-β-alanine (HMDB: 00026) was depleted in brain.
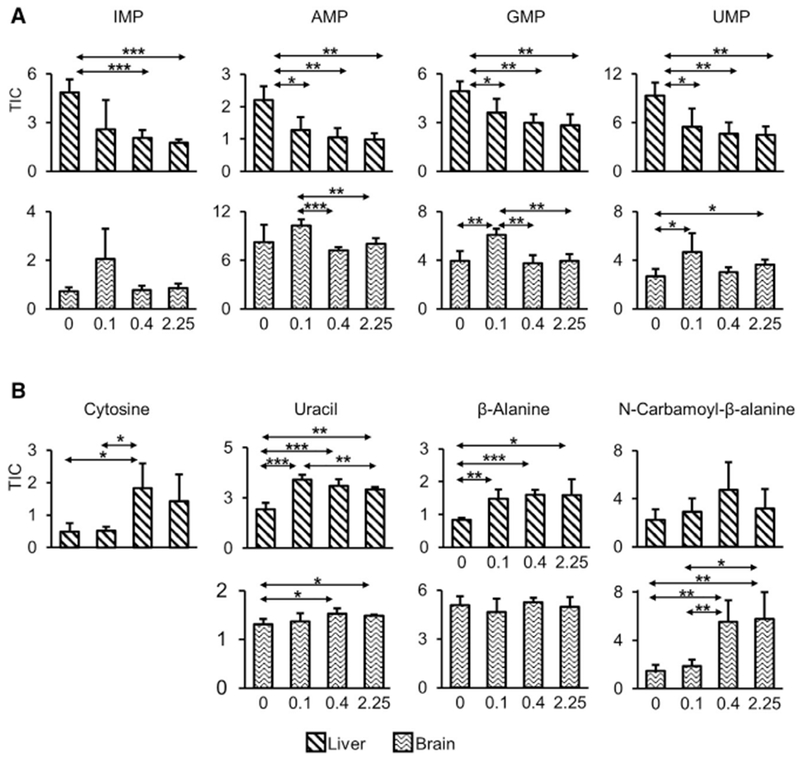
(A) Mononucleotide levels, IMP, AMP, GMP, and UMP in liver (upper panel) and brain (lower panel). All four mononucleotides measured in the liver were increased by Se deficiency.
(B) The levels of metabolites involved in pyrimidine catabolic pathways were decreased by Se deficiency in an organ-specific manner. Upper graphs show liver and lower graphs show brain. TIC represents total ion current.
*p < 0.05, **p < 0.01, and ***p < 0.001 (two-tailed Student’s t test).
Se deficiency altered 63 (0 versus 0.1 ppm) and 76 (0 versus 0.4 ppm) metabolites (p < 0.01) in liver; of which, 25 were common (Figure 5A; Table S1). With less-stringent criteria (p < 0.05), 77 metabolites were found to be commonly altered by Se deficiency, 12 by Se toxicity, and 6 metabolites from both dietary regimens. The Se-deficiency-associated metabolites were associated with amino acid and lipid metabolism (Figure S2A). The levels of L-asymmetric dimethyl arginine (ADMA, HMDB: 01539), oxidized glutathione (HMDB: 03337), L-methionine (HMDB: 00696) were reduced, whereas C18:2 lysophosphatidylethanolamine (LPE; HMDB: 11507), S-adenosylmethionine (HMDB: 01185), and methionine sulfoxide (HMDB: 02005) were increased by Se deficiency. These metabolites are associated with metabolic pathways including cysteine and methionine metabolism. Se toxicity altered 46 metabolites in 0.1 versus 2.25 ppm Se diet comparison and six in 0.4 versus 2.25 ppm; among these, two metabolites were commonly changed (p < 0.01): betaine (HMDB: 00043) and S-adenosylmethionine (HMDB: 01185) (Figure 5B; Table S2).
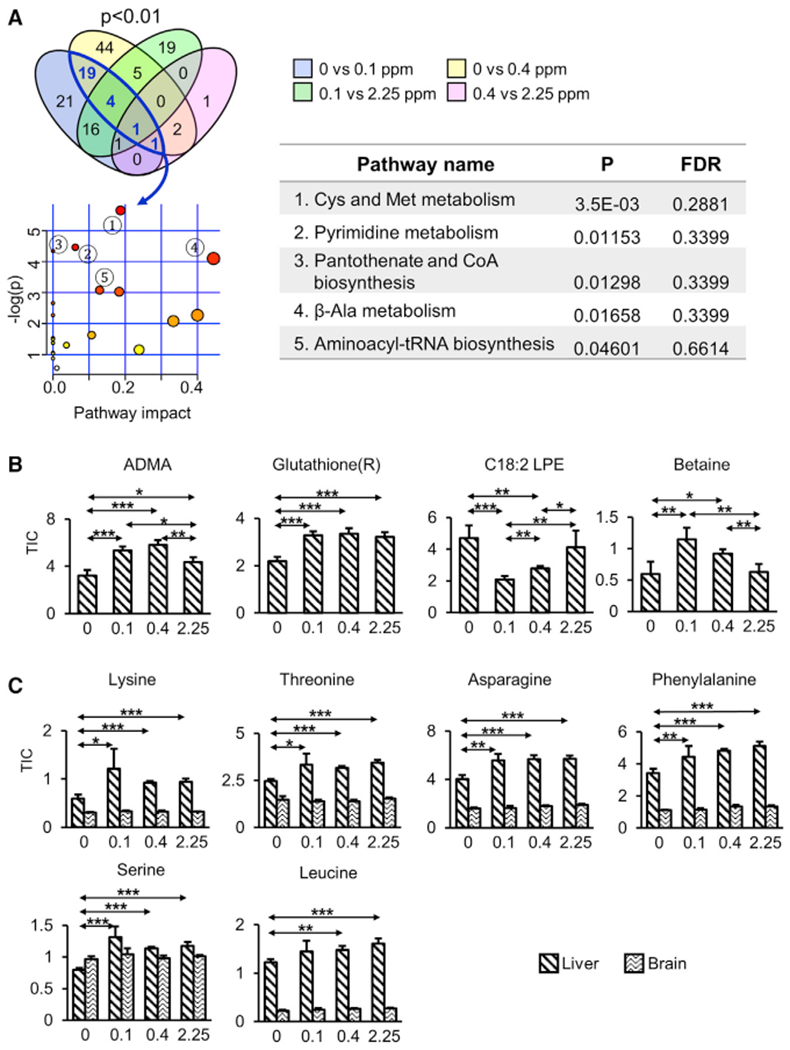
(A) Metabolites associated with Se deficiency (0 versus 0.1 ppm and 0 versus 0.4 ppm) and Se toxicity (0.1 versus 2.25 ppm and 0.4 versus 2.25 ppm) were quantified at p < 0.01. Enriched pathways commonly regulated by Se deficiency were assessed with MetaboAnalyst 4.0 and the top five pathways are shown in the table. The axis titles for metabolome view are shown in Figure 2E.
(B) Examples of metabolites altered under conditions of Se deficiency and Se toxicity.
(C) Depletion offree amino acids in liver in response to Se deficiency. Six amino acids with significantly (p < 0.001) lower levels in Se-deficient liver are shown. Brain data are shown for comparison.
(B and C) *p < 0.05, **p < 0.01, and ***p < 0.001 (two-tailed Student’s t test).
When considered as a whole, the most drastic changes observed were the depletion of amino acids and their derivatives in the liver by Se deficiency (but not in the brain). Our targeted metabolite profiling measured 19 amino acids (cysteine was not analyzed). Asparagine, leucine, lysine, methionine, phenylalanine, serine, and threonine were reduced by Se deficiency (p < 0.001) when compared with 0.1 or 0.4 ppm diets (Figure 5C). Upon depletion of amino acid levels, mTOR is inhibited and, in turn, regulates a host of downstream events that modulate growth and metabolism. Genetic inhibition of mTOR signaling can extend the lifespan in a wide range of organisms, including worms (Vellai et al., 2003), flies (Kapahi et al., 2004), and mice (Lamming et al., 2012; Wu et al., 2013).
In brain, Se deficiency altered the levels of three metabolites: inositol (HMDB: 00211), tryptophan (HMDB: 00929), and putrescine (HMDB: 01414), whereas amino acids and their derivatives, polyamines, and methyl biosynthesis-associated metabolites were affected by Se toxicity. Eight metabolites were commonly altered, including those linked with methionine (methionine sulfoxide (HMDB: 02005), 5-adenosylhomocysteine (HMDB: 00939), and methionine (HMDB: 00696)) (Figure S2B).
Regulation of Gene Expression by Dietary Se in Liver and Brain
To link changes in the metabolome with those in the transcriptome, we subjected liver and brain to RNA sequencing (RNA-seq) (Data S2). Principal-component analysis (PCA) showed that samples clustered mainly by the tissue of origin (Figure 6A). Regardless of dietary Se, brain transcriptomes were little affected, in contrast with those in the liver. Among selenoproteins, Selenop (code for a secreted plasma protein that is primarily expressed in liver and delivers Se to other organs) had the greatest number of reads in the liver, and many other selenoprotein mRNAs were also more abundant in this organ. Only Selenom, Selenon, Selenow, GPx3, and Dio2 transcripts were more abundant in the brain than in the liver (Figures 6B and S3). Transcripts that were affected most significantly by Se deficiency in liver (Figure 6C) included selenoproteins GPx1 and Selenow. Differentially expressed transcripts were further examined in more detail in liver and brain (Figure S4). Among the top 100 transcripts affected by Se deficiency, 81 showed less expression compared with controls (Se-sufficient diets). Transcripts with decreased expression in response to Se deficiency were further analyzed for their interaction and pathway enrichment (Figure 6D), revealing “biosynthesis of amino acids” (FDR, 4.27E–06) as the most affected process.
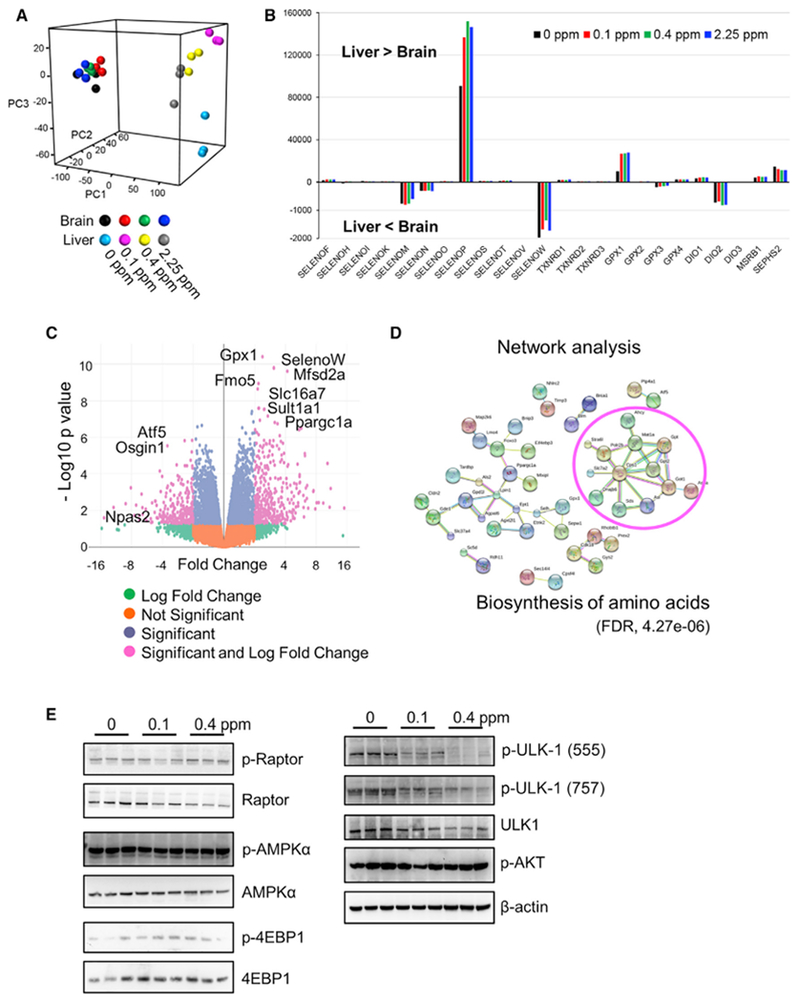
(A) Principal components ofthe global transcriptome of liver and brain of mice fed 0, 0.1, 0.4, or 2.25 ppm Se diets. Each point represents one mouse and color indicates Se dietary groups.
(B) Selenoprotein gene expression in liver and brain. The y axis indicates the total read counts for the indicated transcripts. Positive values indicate that there are more reads in the liver than in the brain, and negative values that there are more reads in the brain than in the liver.
(C) Volcano plot of transcripts affected by Se deficiency (comparison between 0 and 0.1 ppm Se diets). Each dot corresponds to a single transcript, and pink dots are significant in both −log p values (y axis) and fold change (x axis). Right: this corresponds to transcripts decreased by Se deficiency. Left: those increased by Se deficiency.
(D) Network analysis of enriched pathways and interactions. A total of 81 transcripts with lower expression in response to Se deficiency (based on −log p value) were analyzed and visualized with STRING (https://string-db.org). Highly clustered transcripts circled in red correspond to biosynthesis of amino acids (FDR, 4.27E–06).
(E) Western blot analyses of the livers of mice subjected to the indicated Se diets (0, 0.1, 0.4, and 2.25 ppm). Proteins probed are indicated on the right. Ulk1 is highly phosphorylated by Se deficiency.
As deficiency in selenoproteins Txnrd1 and GPx4 is known to activate nuclear factor (erythroid-derived 2)-like 2 (Nrf2), and Se deficiency is also implicated in this pathway (Brigelius-Flohé and Kipp, 2013; Burk et al., 2008; Cebula et al., 2015; Johansson et al., 2017; Locy et al., 2012; Müller et al., 2010; Schmidt, 2015), we were particularly interested in examining the dietary Se-dependent Nrf2 response. Specifically, we tested whether the 0 ppm Se diet affected Nrf2 target genes at the level of transcripts and proteins by analyzing the expression of two of the most well-characterized Nrf2 targets, heme oxygenase-1 (HO-1) and NAD[P]H: quinone oxidoreductase 1 (NQO1), along with Nrf2 itself. However, the liver of mice fed the 0 ppm Se-deficient diet did not modify the expression of Nrf2 target proteins or its own levels (Figure S5). We further compiled the list of known Nrf2 targets and examined Nrf2 target enrichment by gene set enrichment analysis (GSEA) analysis but, again, found no difference in the expression of these genes.
To further interrogate the molecular mechanisms behind Se deficiency, we examined the levels and modification status of regulators associated with AMPK and mTOR pathways. Among these, Ulk1 phosphorylation status was elevated in the Se-deficient liver (Figure 6E). p-Ulk1 is involved in cellular nutrient homeostasis and supports an interplay between mTOR and AMPK pathways. Increased phosphorylation of Ulk1 is consistent with the low amino acid status and decreased expression of genes associated with the biosynthesis of amino acids.
Dietary Se Regulates miRNA Expression in Liver
Another mechanism of regulation by dietary Se may involve microRNA (miRNA) expression. We examined whether miRNA levels are affected by Se, which may, in turn, influence selenoprotein expression by analyzing the expression of more than 600 miRNAs in mouse liver with NanoString technology (Data S3). Ten miRNAs were differently expressed between 0 versus 0.1 ppm Se diets, and 30 were differentially expressed between 0 and 0.4 ppm. However, no miRNA was commonly altered by Se deficiency compared with 0.1 ppm and 0.4 ppm diets. In addition, none of the altered miRNAs were predicted to target multiple selenoprotein transcripts. On the other hand, we identified 22 miRNAs regulated by Se toxicity relative to 0.4 ppm Se and 26 relative to 0.1 ppm Se (Figure S6).
The expression levels of 38 miRNAs were changed by Se deficiency relative to Se sufficiency (0.1 or 0.4 ppm). After mapping them onto MirSystem, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways associated with MapK signaling pathways (1.8174E–19), Wnt signaling (5.2354E–11), mTOR signaling pathway (1.1005E–10), ribosome (3.3890E–09), and electron carrier activity (1.5583E–08) were enriched. This finding, once again, pointed to the effects on amino acid and protein metabolism. miRNAs differentially expressed in each dietary group (p < 0.05) are shown in Table S3. We further analyzed whether the identified miRNAs could regulate selenoprotein expression in primary mouse hepatocytes. Nanoparticle-forming solutions that contained inhibitors to these miRNAs were individually transfected into hepatocytes, and incorporation of 75Se into selenoproteins was evaluated. None of the hairpin inhibitors affected selenoprotein expression (Figure S6). The data suggest that selenoprotein expression is not directly regulated by these miRNAs; instead, miRNA expression is affected by Se deficiency via decreased selenoprotein expression.
DISCUSSION
We examined the systemic effects of dietary Se at the molecular level by a combination of omics approaches. Being a component of several essential oxidore-ductases, Se is viewed as a regulator of redox homeostasis that protects cells from stress and inflammation, but many of its biological functions are incompletely understood. Se deficiency decreases the levels of many of the selenoproteins, compromising a plethora of cellular functions and leading to adverse health conditions. To examine how dietary Se affects molecular function, we subjected mice to Se-deficient (0 ppm), Se-sufficient (0.1 ppm and 0.4 ppm), and toxic Se (2.25 ppm) diets. Our analyses confirmed that Se deficiency dramatically reduced selenoprotein expression, whereas, as expected, selenoprotein expression was little affected when the Se levels were increased above Se sufficiency. We also observed the expected priority for Se supply in different organs: liver was easily depleted of selenoproteins in response to Se deficiency, whereas brain selenoprotein expression was essentially unchanged.
Systemic and long-term effects of interventions may be reflected in such integrative parameters as the rate of aging. Remarkably, Se-deficient diets given mice for the entire adult life did not shorten their lifespan. This was surprising considering the requirement of dietary Se for so many proteins involved in redox homeostasis and other metabolic pathways, as well as considering the well-established detrimental effects of dietary Se deficiency. Although our investigation is the first study to our knowledge to examine the effect of dietary Se in mice with a normal lifespan, our finding is consistent with the recent observation of the increased lifespan of short-lived G3 Terc−/− mice subjected to Se deficiency (Wu et al., 2017). In addition, GPx4 heterozygous knockout mice showed an increased lifespan and reduced age-related pathologies (Ran et al., 2007). Se has long been implicated in aging, and the fact that even such a drastic treatment as extreme Se deficiency did not reduce lifespan points to molecular mechanisms that alleviate the consequences of Se deficiency. It should be noted that our experiments were performed in a laboratory setting, so the findings may not necessarily be applicable to animals in the wild. However, these findings may be relevant to humans because their mortality patterns resemble those of animals in protected environments.
To identify the molecular changes associated with lifespan control under conditions of Se deficiency, we performed metabolite profiling, RNA-seq, and miRNA expression analyses. Interestingly, Se deficiency affected metabolite levels and gene expression more drastically than other diets did, e.g., it affected more metabolites than the 2.25 ppm diet (compared with the 0.1- and 0.4 ppm Se diets). Thus, it is likely that Se deficiency influenced the metabolome through reduced expression of selenoproteins. Another observation was that dietary Se largely targeted the metabolome and transcriptome in liver and less so in the brain. The key finding in our targeted metabolite profiling was that dietary Se changed the levels of amino acids and mononucleotides. Among the 19 measured amino acids, seven (asparagine, leucine, lysine, methionine, phenylalanine, serine, and threonine) were decreased (p < 0.001) in response to Se deficiency in liver, whereas several mononucleotides (IMP, AMP, GMP, and UMP) increased. Both amino acid and mononucleotide levels are known to influence the aging process through nutrient sensing and energy homeostasis. As an example, methionine restriction extends the lifespan of rats and mice and improves glucose handling (Ables et al., 2012; Lees et al., 2017; Malloy et al., 2006; Malloy et al., 2013; Miller et al., 2005; Richie et al., 1994; Stone et al., 2014; Wanders et al., 2015). Amino acid levels regulate the activity of mTOR, whose inhibition leads to lifespan extension, whereas higher levels of lower-energy adenine nucleotides, such as AMP, indicate lower cellular energy stores that can activate AMPK, similar to the effects of dietary restriction. Thus, metabolome remodeling in response to Se deficiency may be viewed as a mimic of calorie restriction in its influence on longevity pathways.
Among amino acids, serine was decreased the most by Se deficiency. This amino acid provides the backbone for the synthesis of Sec, suggesting another mechanism (in addition to Se availability) regulates selenoprotein synthesis. Serine and glycine are also precursors for the synthesis of proteins, nucleic acids, and lipids. In many cancer cells, serine biosynthesis represents a critical metabolic junction; this amino acid is also thought to be the third most consumed metabolite (in addition to glucose and glutamine) (Frezza, 2016). Amino acids represent most of the carbon in mammalian cells, with the serine carbons incorporated into proteins, nucleic acids, creatine, porphyrins, phospholipids, and glutathione (Labuschagne et al., 2014; Locasale and Cantley, 2011; Possemato et al., 2011). In addition, serine, glycine, and other one-carbon metabolism components have roles in folate and methionine cycles, and upregulation of these pathways was also associated with cancer pathogenesis (Locasale, 2013).
Together with cysteine, methionine is a sulfur-containing proteinogenic amino acid; it can be reversibly oxidized and can influence protein function (Banerjee, 2012). We found that methionine metabolism was also altered by Se deficiency, consistent with the idea that this dietary regimen leads to oxidative stress (a subset of metabolic pathways targeted by Se are summarized in Figure 3B). It was also shown that a metabolic pathway that uses glutathione derived from methionine can sustain reductive pathways in the mouse lacking both selenoprotein Txnrd1 and glutathione reductase (GR) in the liver (Eriksson et al., 2015; Prigge et al., 2017). In addition to regulation of redox homeostasis, Se deficiency is known to decrease global DNA methylation (Davis et al., 2000). Interestingly, SAM in liver was 3-fold greater in the Se-deficient group, but its levels were not altered in brain. Choline, betaine, and dimethylglycine, which belong to the homocysteine salvage pathway were also decreased significantly by dietary Se deficiency. Betaine is an import resource of one-carbon units under conditions of folate deficiency; it functions as an organic osmolyte and is involved in methylation reactions and detoxification of homocysteine. The homocysteine salvage pathway was significantly decreased in response to Se deficiency, suggesting that methylation to facilitate necessary chemical processes, cellular replication, and detoxification may also be attenuated. We further found that ADMA was lower in both the 0 and 2.25 ppm groups. ADMA is an endogenous inhibitor of nitric oxide synthase at concentrations found in pathophysiological conditions. Hypercholesterolemia, atherosclerosis, hypertension, chronic renal failure, and chronic heart failure potentiate ADMA in the human plasma (Böger, 2003). Our metabolite profiling data were consistent with the idea that altered ADMA levels may be associated with a lower risk of endothelial dysfunction and coronary artery disease. Overall, metabolite profiling revealed significant changes in metabolites in response to dietary Se, most notably, changes in amino acids, mononucleotides, redox homeostasis, and methionine metabolism. Attenuation of amino acid levels in response to Se deficiency was a particularly interesting observation, linking dietary Se with a complex web of metabolic pathways associated with insulin resistance, diabetes, cardiac and skeletal muscle biogenesis, and lifespan control (D’Antona et al., 2010; Grandison et al., 2009).
Additional evidence for the observed regulatory mechanisms came from RNA-seq data, which we performed to investigate how dietary Se affected gene expression in mouse organs. Consistent with the metabolome analyses, we observed lower variance in gene expression in brain than we did in liver. It is known that brain has a higher priority for Se supplies and that, even under conditions of Se deficiency, it largely maintains selenoprotein expression, whereas liver may lose more than 90% of its Se and abundant selenoproteins, such as GPx1. Indeed, GPx1 and Selenow transcripts were the most affected by Se deficiency in the liver, and several other selenoprotein transcripts also decreased significantly.
To gain further insights into the processes that underlie responses to Se deficiency, we analyzed differentially expressed genes and found that the most overrepresented pathway was “biosynthesis of amino acids” (FDR, 4.27E–06), which was downregulated. This data further support the idea that Se deficiency decreases cellular amino acids through nutrient-sensing mechanisms. Nutrient sensing is known to mediate the beneficial effects on longevity. Interconnected nutrient-sensing systems include regulators, such as mTOR, for sensing cellular amino acid levels, AMPK that senses low-energy states by detecting high AMP levels and sirtuins that sense high nicotinamide adenine dinucleotide (NAD+) levels of low-energy states. Interestingly, we found that Ulk1 phosphorylation was increased by Se deficiency in the liver, further supporting the association between Se deficiency and pro-longevity mechanisms linked to nutrient sensing. It is well established that anabolic signaling accelerates aging, and decreased nutrient signaling extends longevity (Fontana et al., 2010), and Se deficiency fits into this picture, raising the possibility that it represents a pro-longevity treatment.
Previous studies implicated the deficiency of two selenoproteins, Txnrdl and GPx4, as well as dietary Se deficiency in activation of transcription factor Nrf2 and the consequent expression of a battery of maintenance genes (Burk et al., 2008; Dong et al., 2016; Mostert et al., 2003; Müller et al., 2010). However, we found that the levels of Nrf2 target proteins HO-1 and NQO1 as well as their transcripts were not affected by Se deficiency. This was recapitulated by the analysis of whole-transcriptome remodeling in response to the 0 ppm Se diet, which revealed no changes in the expression of Nrf2-response genes. How can it be that the knockout of selenoproteins Txnrdl or GPx4 activated Nrf2, whereas general Se deficiency in our study did not? Moreover, some previous studies found that Se deficiency may activate HO-1 and NQO1 (Burk et al., 2008). In the antioxidant-response element (ARE)-reporter mice, this antioxidant element could also be activated by Se deficiency (Mostert et al., 2003). However, another study found that Se deficiency affected the expression of neither HO-1 nor NQO1 (Dong et al., 2016). However, in that same study Se deficiency dramatically decreased hepatic Se levels, GPx1 activity and expression, Txnrd1 activity and expression, and serum GPx activity. In addition, injection of EGCG (epigallocatechin gallate, a control for Nrf2 activation) to either Se-adequate or Se-deficient mice could induce HO-1. Thus, in that study, similar to our study, Se deficiency decreased the expression of important selenoproteins but did not activate Nrf2. In addition, Se deficiency affected selenoprotein expression, but not Nrf2 targets, in a study that examined gene expression and translation (by ribosome profiling) (Tsuji et al., 2015). Thus, the data suggest that Se deficiency interacts with another dietary or environmental component to regulate the Nrf2 response, but, by itself, it is not sufficient. Clearly, several studies, including ours, observed robust selenoprotein deficiency (measured by the expression of GPx1, hepatic Se levels, etc.) yet no Nrf2 activation. Although Nrf2 can be activated by the knockout of Txnrd1 and GPx4, under conditions of Se deficiency, these proteins are only partially affected with regard to expression levels. Thus, Se deficiency and Nrf2 activation are uncoupled, i.e., Se deficiency is not sufficient for activation of Nrf2 but could activate it either together with other environmental factor or under conditions of knockout of certain selenoproteins.
Additional insights were provided by the analysis of miRNA expression. Several miRNAs altered in response to dietary Se in mouse liver. As Se-dependent changes in the metabolome and gene expression were dramatic, such miRNA responses likely included both direct and indirect effects. A possibility that the identified miRNAs directly affect selenoprotein expression was then tested. However, their inhibition yielded no changes in 75Se incorporation into selenoproteins, arguing against the direct effect. Further analyses are needed to delineate the relationship between selenoprotein expression and miRNA expression, but our data suggest that selenoproteins as a class are not commonly regulated by miRNAs (this does not exclude regulation in a selenoprotein-specific manner) and that miRNA expression is regulated indirectly, likely via altered expression of selenoproteins.
Taken together, our findings show that long-term Se deficiency in adult mice is compatible with a normal lifespan and triggers metabolic perturbations and signaling effects reminiscent of nutrient sensing. As such, Se deficiency emerges as a prolongevity condition. It would be interesting to examine in more detail how this process interacts with aging. Age-associated decline of protein synthesis can be attenuated by dietary restriction in mice and rats, and lifespan may be increased by such dietary regiments as calorie restriction and methionine restriction (Harrison etal., 2009; Neff etal., 2013). During the aging process, muscle loss may affect the total body pool of Se and its availability as skeletal muscle and liver appear to act as major storage organs for this element (Oster et al., 1988). Clearly, further studies are warranted to delineate the pro-longevity states induced by dietary Se deficiency. At least within a certain window of concentration, Se deficiency may lead to an increase in lifespan (Wu et al., 2017). Such an effect is also consistent with the reduced use of Se in long-lived mammals and the partial loss of Sec in selenoproteins in these organisms (Kasaikina et al., 2011; Lobanov et al., 2008).
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Vadim N. Gladyshev (ude.dravrah.hwb.scir@vehsydalgv).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals and feeding protocols
Rodent chow diets containing 0,0.1,0.4, or 2.25 μg/g (ppm) Se as sodium selenite were custom manufactured by Harland TekLad based on the Torula yeast-based diet (TD.92163) (Madison, WI). The 0 ppm Se diet was considered Se-deficient, the 0.1 ppm corresponded approximately to the lower end of normal GPx1 expression, which is maximized by 0.15 ppm Se. The 0.4 ppm Se corresponded to an approximate level of Se in human clinical trials, and 2.25 ppm Se represented enriched Se levels approaching Se toxicity.
To investigate how dietary Se affects lifespan, 342 weaning mice were subjected to 0 ppm, 0.1 ppm, and 0.4 ppm Se diets. Males and females were randomly assigned to one of three experimental groups for a total of 57 mice/group·sex, allowed ad libitum access to the designated diet, and followed up to 28.5 months when the study was terminated.
To perform metabolite profiling and miRNA expression analysis, short-term Se diet regime was applied. Eight-week-old C57BL/6J male mice were obtained (Jackson Laboratory, Bar Harbor, ME) and fed with 0, 0.1, 0.4, and 2.25 ppm Se diets for 5 weeks (5 mice per group). At the end of the study, mice were sacrificed, tissues were removed, snap frozen, and stored at −80°C until further analysis. Experimental animal protocols were approved by the Institutional Animal Care and Use Committee and were performed according to NIH guidelines.
METHOD DETAILS
Metabolite profiling
Metabolite profiling was performed on mouse liver and brain tissue samples that were harvested from mice subjected to for five weeks to 0, 0.1, 0.4, 2.25 ppm Se diets. Tissue metabolites were analyzed using liquid chromatography-tandem mass spectrometry (LC-MS) with high resolution accurate mass profiling. All data were acquired using LC-MS systems comprised of a Nexera X2 U-HPLC (Shimadzu, Marlborough, MA) coupled to either a Q Exactive or Exactive Plus orbitrap mass spectrometer (Thermo Fisher Scientific; Waltham, MA) (Avanesov et al., 2014; Ma et al., 2015). Raw data from the three nontargeted methods were processed using Progenesis QI software (NonLinear Dynamics) for feature alignment, nontargeted signal detection, and signal integration. Targeted processing of a subset of known metabolites was conducted using TraceFinder 3.3 software (Thermo Fisher Scientific). Compound identities were confirmed using reference standards and reference samples.
RNA sequencing
RNaseq libraries (three mice per dietary group) were prepared with Illumina TruSeq RNA kits (poly A Selection). Sequencing was performed using the latest HiSeq Illumina Sequencing Systems with 100 bp paired-end reads to obtain minimum 3Gb throughput data. Raw RNaseq fastq reads were aligned to mouse genome (mm10) using STAR (v2.5) (Dobin and Gingeras, 2016). Genes were sub-sequently counted using Rsubread and analyzed for gene expression changes using limma-voom with quantile normalization (Ritchie et al., 2015). The gene- and sample-specific normalization factors were then used to correct counts to generate bedgraph files used for further visualization. The pathway enrichment was tested with STRING (https://string-db.org) and Gene Set Enrichment Analysis (GSEA) (Subramanian et al., 2007; Subramanian et al., 2005).
MicroRNA analysis
microRNA expression profiling was performed using the NanoString nCounter® microRNA Expression Assays kit (NanoString, Seattle, WA) according to manufacturer’s protocol. Each microRNA is identified by the “color code” generated by six ordered fluorescent spots in the reporter probe. Briefly, the total RNA was isolated from the snap frozen livers of individual mouse using TRIzol® Reagent (Thermo Fisher scientific) and cleaned up with QIAGEN RNA purification kit. The NanoString mouse microRNA panel detects more than 600 murine and murine-associated viral microRNAs (miRBase data base; http://www.mirbase.org). The counted and tabulated raw data for each microRNA species were then normalized to internal positive spike controls. The average of the three biological replicates was calculated and used for comparison with expression profiles obtained from each Se diet. The microRNA target prediction and pathway mapping were performed with Targetscan (http://www.targetscan.org/), KEGG pathway (https://www.genome.jp/kegg/pathway.html) mapping, MirSystem (http://mirsystem.cgm.ntu.edu.tw/index.php), and Diana tools (http://diana.imis.athena-innovation.gr/DianaTools/index.php, mirPath v. 3, Tarbase and microT-CDS).
Primary hepatocyte culture, transfections of miRNA hairpin inhibitors using N-TER™ nanoparticles, and labeling cells with 75Se
Mouse primary hepatocytes were isolated from 8-week old C57BL/6J wild-type mice (Tanaka et al., 2015). Isolated hepatocytes were further washed with 0.1 M phosphate-buffered saline (PBS) and purified with Percoll gradient (GE Healthcare), counted, and seeded in collagen coated 6 well plates at a density of 3 × 105 cells/well. Primary hepatocytes were attached in RPMI 1640 medium (Lonza, Switzerland) with 10% fetal bovine serum, 2 mM glutamine, and antibiotics for 6 h. For transfection of miRNA inhibitors, 20 custom made Hairpin Inhibitors for indicated mouse miRNAs were purchased (GE Dharmacon) and transfected into hepatocytes using N-TER™ (Sigma). Briefly, nanoparticle forming solutions (NFS) containing miRIDIAN microRNA Hairpin inhibitors were prepared by following the N-TER™ peptide protocol (Sigma). Metabolic labeling of primary hepatocytes with 75Se was performed by the addition of freshly neutralized 75Se to cells (5 μCi/mL of medium) and incubated for 24 h at 37°C in a humidified incubator. After labeling of cells with 75Se for 24 h, the cells were harvested, lysed, proteins quantified, and separated on SDS-PAGE. The separated proteins were transferred onto PVDF membranes and analyzed with a PhosphorImager (Yim et al., 2018).
QUANTIFICATION AND STATISTICAL ANALYSIS
Unless otherwise stated, two tailed Student’s t test was used to calculate the differences between Se dietary groups. Error bars shown in the figures indicate standard deviation of the mean, and all p values or asterisk marks represent statistical significance (***p < 0.001, **p < 0.01, and *p < 0.05). For longevity studies, survival curves were generated, and the groups were compared using the log-rank test using STATA statistical program to evaluate the difference.
DATA AND SOFTWARE AVAILABILITY
All primary LC-MS metabolite profiling data, RNaseq result, miRNA data are available as supplemental files. Raw RNA-seq data reported in this paper were submitted to the Gene Expression Omnibus database available at https://www.ncbi.nlm.nih.gov/geo/ under the accession code {"type":"entrez-geo","attrs":{"text":"GSE129138","term_id":"129138","extlink":"1"}}GSE129138.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-GPxl | ABfrontier (Seoul, Korea) | LF-PA0235 |
| anti-β-Actin | Sigma | Monoclonal AC-74 (A2228) |
| anti-MsrB1 | Kim and Gladyshev, 2004; Turanov et al., 2010 | Clone ID: NE 111 |
| anti-TR1 (Txnrd1) | Kim and Gladyshev, 2004; Turanov et al., 2010 | Clone ID:165 |
| anti-TR3 (Txnrd2) | Kim and Gladyshev, 2004; Turanov et al., 2010 | Clone ID:157 |
| anti-Trx2 | Kim and Gladyshev, 2004; Turanov et al., 2010 | Clone ID:173 |
| anti-HO-1 | Novus Biologicals | NBP1-31341 |
| anti-NQO1 | Novus Biologicals | NB-200-209 |
| anti-Nrf2 | Abcam | ab137550 |
| anti-p-ULK1 | Cell Signaling Technology, sampler kit (8359T) | Ser 555 (mAb 5869) Ser 757 (mAb 6888) |
| anti-ULK1 | Cell Signaling Technology, sampler kit (8359T) | D8H5 (mAb 8054) |
| anti-p-Raptor | Cell Signaling Technology, sampler kit (8359T) | Ser 792 (mAb 2083) |
| anti-Raptor | Cell Signaling Technology, sampler kit (8359T) | 24C12 (mAb 2280) |
| anti-p-AMPKα | Cell Signaling Technology, sampler kit (8359T) | Thr 172 (mAb 2535) |
| anti-AMPKα | Cell Signaling Technology, sampler kit (8359T) | D63G4 (mAb 5832) |
| anti-p-4EBP1 | Cell Signaling Technology Phospho-4E-BP1 (Thr70) Antibody | Thr 70 (mAb 9455) |
| anti-4EBP1 | Cell Signaling Technology 4E-BP1 Antibody | mAb 9452 |
| anti-p-AKT | Cell Signaling Technology Phospho-Akt (Ser 473) Antibody | Ser 473 (mAb 9271) |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Percoll | GE Healthcare | 17089101 |
| miRNA inhibitors (miRIDIAN) | GE Dharmacon | Custom designed (the microRNA inhibitors are in Figure S6B) |
| N-TER™ | Sigma | N2913 |
| TRIzol® Reagent | Thermo Fisher scientific | 15596026 |
| Critical Commercial Assays | ||
| NanoString nCounter® microRNA Expression Assays kit | NanoString | CSO-MMIR15-12 |
| QIAGEN RNA purification kit | QIAGEN | 74104 |
| TruSeq RNA kits | Illumina | TruSeq RNA Library Prep Kit v2 |
| Deposited Data | ||
| Raw sequencing data for RNaseq | This paper (GEO dataset) https://www.ncbi.nlm.nih.gov/gds/?term={"type":"entrez-geo","attrs":{"text":"GSE129138","term_id":"129138"}}GSE129138 | {"type":"entrez-geo","attrs":{"text":"GSE129138","term_id":"129138","extlink":"1"}}GSE129138 |
| Experimental Models: Organisms/Strains | ||
| Mouse (C57BL/6J) | Jackson Laboratory | 000664 |
| Mouse (BALB/cJ) | Jackson Laboratory | 000651 |
| Software and Algorithms | ||
| MetaboAnalyst 4.0 | https://www.metaboanalyst.ca/faces/home.xhtml | Chong et al., 2018 |
| STAR (v2.5) RNA-seq aligner | https://github.com/alexdobin/STAR | STAR 2.7 |
| STRING | https://string-db.org | Version 11.0 |
| Gene Set Enrichment Analysis (GSEA) | http://software.broadinstitute.org/gsea/index.jsp | Subramanian et al., 2007; Subramanian et al., 2005 |
| Other | ||
| Rodent Selenium Diet (0, 0.1, 0.4, or 2.25 μg/g (ppm) Selenium containing diet) | Harland TekLad | TD.92163 |
| Radioisotope (75Se) | MURR (University of Missouri Research Reactor Center) Columbia, MO | https://www.ehs.missouri.edu/ehs/sites/ehs/files/pdf/isotopedata/se-75.pdf |
Highlights
- Se deficiency reduces selenoprotein expression but does not affect lifespan of mice
- Se deficiency reduces amino acid levels and elevates mononucleotides
- Se deficiency activates pathways linked to nutrient sensing
- Se deficiency is associated with pro-longevity mechanisms
Supplementary Material
1
2
3
4
ACKNOWLEDGMENTS
We thank Sergey Novoselov for carrying out animal lifespan studies, Judith Manola for supporting survival analyses, and Alexei Lobanov, Xuming Zhou, and Marco Mariotti for RNA-seq analyses. This work was supported by NIH GM065204, CA080946, and AG021518 to V.N.G. and GM109312 to S.H.Y. The funding agency had no involvement in study design or data collection and interpretation.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.05.001.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Ables GP, Perrone CE, Orentreich D, and Orentreich N (2012). Methionine-restricted C57BL/6J miceare resistanttodiet-induced obesityand insulin resistance but have low bone density. PLoS ONE 7, e51357. [PMC free article] [PubMed] [Google Scholar]
- Allen NE, Appleby PN, Roddam AW, Tjønneland A, Johnsen NF, Overvad K, Boeing H, Weikert S, Kaaks R, Linseisen J, et al.; European Prospective Investigation into Cancer and Nutrition (2008). Plasma selenium concentration and prostatecancer risk: resultsfrom the European Prospective Investigation into Cancer and Nutrition (EPIC). Am. J. Clin. Nutr 88, 1567–1575. [PubMed] [Google Scholar]
- Anan Y, and Ogra Y (2013). Toxicological and pharmacological analysis of selenohomolanthionine in mice. Toxicol. Res. (Camb.) 2, 115–122. [Google Scholar]
- Arnaudguilhem C, Bierla K, Ouerdane L, Preud’homme H, Yiannikouris A, and Lobinski R (2012). Selenium metabolomics in yeast using complementary reversed-phase/hydrophilic ion interaction (HILIC) liquid chromatography-electrospray hybrid quadrupole trap/Orbitrap mass spectrometry. Anal. Chim. Acta 757, 26–38. [PubMed] [Google Scholar]
- Arnér ES (2009). Focus on mammalian thioredoxin reductases—important selenoproteins with versatile functions. Biochim. Biophys. Acta 1790, 495–526. [PubMed] [Google Scholar]
- Avanesov AS, Ma S, Pierce KA, Yim SH, Lee BC, Clish CB, and Gladyshev VN (2014). Age- and diet-associated metabolome remodeling characterizes the aging process driven by damage accumulation. eLife 3, e02077. [PMC free article] [PubMed] [Google Scholar]
- Banerjee R (2012). Redox outside the box: linking extracellular redox remodeling with intracellular redox metabolism. J. Biol. Chem 287, 4397–4402. [PMC free article] [PubMed] [Google Scholar]
- Barnes KM, Evenson JK, Raines AM, and Sunde RA (2009). Transcript analysis of the selenoproteome indicates that dietary selenium requirements of rats based on selenium-regulated selenoprotein mRNA levels are uniformly less than those based on glutathione peroxidase activity. J. Nutr 139, 199–206. [PMC free article] [PubMed] [Google Scholar]
- Bodnár D, Ruzsnavszky O, Oláh T, Dienes B, Balatoni I, Ungvári É, Benkő I, Babka B, Prokisch J, Csernoch L, and Szentesi P (2016). Dietary selenium augments sarcoplasmic calcium release and mechanical performance in mice. Nutr. Metab. (Lond.) 13, 76. [PMC free article] [PubMed] [Google Scholar]
- Böger RH (2003). The emerging role of asymmetric dimethylarginine as a novel cardiovascular risk factor. Cardiovasc. Res 59, 824–833. [PubMed] [Google Scholar]
- Brigelius-Flohé R, and Kipp AP (2013). Selenium in the redox regulation of the Nrf2 and the Wnt pathway. Methods Enzymol. 527, 65–86. [PubMed] [Google Scholar]
- Burk RF, Hill KE, Nakayama A, Mostert V, Levander XA, Motley AK, Johnson DA, Johnson JA, Freeman ML, and Austin LM (2008). Selenium deficiency activates mouse liver Nrf2-ARE but vitamin E deficiency does not. Free Radic. Biol. Med 44, 1617–1623. [PMC free article] [PubMed] [Google Scholar]
- Cebula M, Schmidt EE, and Arner ES (2015). TrxR1 as a potent regulator of the Nrf2-Keap1 response system. Antioxid. Redox Signal 23, 823–853. [PMC free article] [PubMed] [Google Scholar]
- Chong J, Soufan O, Li C, Caraus I, Li S, Bourque G, Wishart DS, and Xia J (2018). MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 46 (W1), W486–W494. [PMC free article] [PubMed] [Google Scholar]
- Christen WG, Glynn RJ, Gaziano JM, Darke AK, Crowley JJ, Goodman PJ, Lippman SM, Lad TE, Bearden JD, Goodman GE, et al. (2015). Age-related cataract in men in the selenium and vitamin e cancer prevention trial eye endpoints study: a randomized clinical trial. JAMA Ophthalmol. 133, 17–24. [PMC free article] [PubMed] [Google Scholar]
- D’Antona G, Ragni M, Cardile A, Tedesco L, Dossena M, Bruttini F, Caliaro F, Corsetti G, Bottinelli R, Carruba MO, et al. (2010). Branched-chain amino acid supplementation promotes survival and supports cardiac and skeletal muscle mitochondrial biogenesis in middle-aged mice. Cell Metab. 12, 362–372. [PubMed] [Google Scholar]
- Davis CD, Uthus EO, and Finley JW (2000). Dietary selenium and arsenic affect DNA methylation in vitro in Caco-2 cells and in vivo in rat liver and colon. J. Nutr 130, 2903–2909. [PubMed] [Google Scholar]
- de Cabo R, Carmona-Gutierrez D, Bernier M, Hall MN, and Madeo F (2014). The search for antiaging interventions: from elixirs to fasting regimens. Cell 157, 1515–1526. [PMC free article] [PubMed] [Google Scholar]
- Dennert G, Zwahlen M, Brinkman M, Vinceti M, Zeegers MP, and Horneber M (2011). Selenium for preventing cancer. Cochrane Database Syst. Rev (5), CD005195. [PMC free article] [PubMed] [Google Scholar]
- Dobin A, and Gingeras TR (2016). Optimizing RNA-Seq mapping with STAR. Methods Mol. Biol 1415, 245–262. [PubMed] [Google Scholar]
- Dong R, Wang D, Wang X, Zhang K, Chen P, Yang CS, and Zhang J (2016). Epigallocatechin-3-gallate enhances key enzymatic activities of hepatic thioredoxin and glutathione systems in selenium-optimal mice but activates hepatic Nrf2 responses in selenium-deficient mice. Redox Biol. 10, 221–232. [PMC free article] [PubMed] [Google Scholar]
- Erdman JW, MacDonald IA, and Zeisel SH (2012). Present Knowledge in Nutrition (Wiley; ). [Google Scholar]
- Eriksson S, Prigge JR, Talago EA, Arner ES, and Schmidt EE (2015). Dietary methionine can sustain cytosolic redox homeostasis inthe mouse liver. Nat. Commun 6, 6479. [PMC free article] [PubMed] [Google Scholar]
- Fontana L, Partridge L, and Longo VD (2010). Extending healthy life span—from yeast to humans. Science 328, 321–326. [PMC free article] [PubMed] [Google Scholar]
- Frezza C (2016). Cancer metabolism: addicted to serine. Nat. Chem. Biol 12, 389–390. [PubMed] [Google Scholar]
- Grandison RC, Piper MD, and Partridge L (2009). Amino-acid imbalance explains extension of lifespan by dietary restriction in Drosophila. Nature 462, 1061–1064. [PMC free article] [PubMed] [Google Scholar]
- Hardie DG (2011). AMP-activated protein kinase: an energy sensorthat regulates all aspects of cell function. Genes Dev. 25, 1895–1908. [PMC free article] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, et al. (2009). Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 460, 392–395. [PMC free article] [PubMed] [Google Scholar]
- Hatfield DL, Yoo MH, Carlson BA, and Gladyshev VN (2009). Selenoproteins that function in cancer prevention and promotion. Biochim. Biophys. Acta 1790, 1541–1545. [PMC free article] [PubMed] [Google Scholar]
- Hellwege JN, Palmer ND, Ziegler JT, Langefeld CD, Lorenzo C, Norris JM, Takamura T, and Bowden DW (2014). Genetic variants in selenoprotein P plasma 1 gene (SEPP1) are associated with fasting insulin and first phase insulin response in Hispanics. Gene 534, 33–39. [PMC free article] [PubMed] [Google Scholar]
- Hill KE, Zhou J, McMahan WJ, Motley AK, Atkins JF, Gesteland RF, and Burk RF (2003). Deletion of selenoprotein P alters distribution of selenium in the mouse. J. Biol. Chem 278, 13640–13646. [PubMed] [Google Scholar]
- Hill KE, Wu S, Motley AK, Stevenson TD, Winfrey VP, Capecchi MR, Atkins JF, and Burk RF (2012). Production of selenoprotein P (Sepp1) by hepatocytes is central to selenium homeostasis. J. Biol. Chem 287, 40414–40424. [PMC free article] [PubMed] [Google Scholar]
- Hrdina J, Banning A, Kipp A, Loh G, Blaut M, and Brigelius-Flohé R (2009). The gastrointestinal microbiota affects the selenium status and selenoprotein expression in mice. J. Nutr. Biochem 20, 638–648. [PubMed] [Google Scholar]
- Hurst R, Armah CN, Dainty JR, Hart DJ, Teucher B, Goldson AJ, Broadley MR, Motley AK, and Fairweather-Tait SJ (2010). Establishing optimal selenium status: results of a randomized, double-blind, placebo-controlled trial. Am. J. Clin. Nutr 91, 923–931. [PMC free article] [PubMed] [Google Scholar]
- Institute of Medicine (2000). Dietary reference intakes forvitamin C, vitamin E, selenium, and carotenoids (National Academies Press; ). [Google Scholar]
- Jakupoglu C, Przemeck GK, Schneider M, Moreno SG, Mayr N, Hatzopoulos AK, de Angelis MH, Wurst W, Bornkamm GW, Brielmeier M, and Conrad M (2005). Cytoplasmic thioredoxin reductase is essential for embryogenesis but dispensable for cardiac development. Mol. Cell. Biol 25, 1980–1988. [PMC free article] [PubMed] [Google Scholar]
- Johansson K, Cebula M, Rengby O, Dreij K, Carlstrom KE, Sigmundsson K, Piehl F, and Arner ES (2017). Cross talk in HEK293 cells between Nrf2, HIF, and NF-κB activities upon challengeswith redoxtherapeutics characterized with single-cell resolution. Antioxid. Redox Signal 26, 229–246. [PMC free article] [PubMed] [Google Scholar]
- Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, and Benzer S (2004). Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr. Biol 14, 885–890. [PMC free article] [PubMed] [Google Scholar]
- Kasaikina MV, Lobanov AV, Malinouski MY, Lee BC, Seravalli J, Fomenko DE, Turanov AA, Finney L, Vogt S, Park TJ, et al. (2011). Reduced utilization of selenium by naked mole rats due to a specific defect in GPx1 expression. J. Biol. Chem 286, 17005–17014. [PMC free article] [PubMed] [Google Scholar]
- Kim HY, and Gladyshev VN (2004). Methionine sulfoxide reduction in mammals: characterization of methionine-R-sulfoxide reductases. Mol. Biol. Cell 15, 1055–1064. [PMC free article] [PubMed] [Google Scholar]
- Klein EA, Thompson IM Jr., Tangen CM, Crowley JJ, Lucia MS, Goodman PJ, Minasian LM, Ford LG, Parnes HL, Gaziano JM, et al. (2011). Vitamin E and the risk of prostate cancer: the Selenium and Vitamin E Cancer Prevention Trial (SELECT). JAMA 306, 1549–1556. [PMC free article] [PubMed] [Google Scholar]
- Kristal AR, Till C, Song X, Tangen CM, Goodman PJ, Neuhauser ML, Schenk JM, Thompson IM, Meyskens FL Jr., Goodman GE, et al. (2014). Plasma vitamin D and prostate cancer risk: results from the Selenium and Vitamin E Cancer Prevention Trial. Cancer Epidemiol. Biomarkers Prev 23, 1494–1504. [PMC free article] [PubMed] [Google Scholar]
- Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, Guigó R, and Gladyshev VN (2003). Characterization of mammalian seleno-proteomes. Science 300, 1439–1443. [PubMed] [Google Scholar]
- Labuschagne CF, van den Broek NJ, Mackay GM, Vousden KH, and Maddocks OD (2014). Serine, but not glycine, supports one-carbon meta-bolism and proliferation of cancer cells. Cell Rep. 7, 1248–1258. [PubMed] [Google Scholar]
- Lamming DW, Ye L, Katajisto P, Goncalves MD, Saitoh M, Stevens DM, Davis JG, Salmon AB, Richardson A, Ahima RS, et al. (2012). Rapamycin-induced insulin resistance is mediated by mTORC2 loss and uncoupled from longevity. Science 335, 1638–1643. [PMC free article] [PubMed] [Google Scholar]
- Lees EK, Banks R, Cook C, Hill S, Morrice N, Grant L, Mody N, and Delibegovic M (2017). Direct comparison of methionine restriction with leucine restriction on the metabolic health of C57BL/6J mice. Sci. Rep 7, 9977. [PMC free article] [PubMed] [Google Scholar]
- Lippman SM, Klein EA, Goodman PJ, Lucia MS, Thompson IM, Ford LG, Parnes HL, Minasian LM, Gaziano JM, Hartline JA, et al. (2009). Effect of selenium and vitamin E on risk of prostate cancer and other cancers: the Selenium and Vitamin E Cancer Prevention Trial (SELECT). JAMA 301, 39–51. [PMC free article] [PubMed] [Google Scholar]
- Lobanov AV, Hatfield DL, and Gladyshev VN (2008). Reduced reliance on the trace element selenium during evolution of mammals. Genome Biol. 9, R62. [PMC free article] [PubMed] [Google Scholar]
- Locasale JW (2013). Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat. Rev. Cancer 13, 572–583. [PMC free article] [PubMed] [Google Scholar]
- Locasale JW, and Cantley LC (2011). Genetic selection for enhanced serine metabolism in cancer development. Cell Cycle 10, 3812–3813. [PubMed] [Google Scholar]
- Locy ML, Rogers LK, Prigge JR, Schmidt EE, Arner ES, and Tipple TE (2012). Thioredoxin reductase inhibition elicits Nrf2-mediated responses in Clara cells: implications for oxidant-induced lung injury. Antioxid. Redox Signal 17, 1407–1416. [PMC free article] [PubMed] [Google Scholar]
- Lu J, and Holmgren A (2009). Selenoproteins. J. Biol. Chem 284, 723–727. [PubMed] [Google Scholar]
- Ma S, Yim SH, Lee SG, Kim EB, Lee SR, Chang KT, Buffenstein R, Lewis KN, Park TJ, Miller RA, et al. (2015). Organization of the mammalian metabolome according to organ function, lineage specialization, and longevity. Cell Metab. 22, 332–343. [PMC free article] [PubMed] [Google Scholar]
- MacFarquhar JK, Broussard DL, Melstrom P, Hutchinson R, Wolkin A, Martin C, Burk RF, Dunn JR, Green AL, Hammond R, et al. (2010). Acute selenium toxicity associated with a dietary supplement. Arch. Intern. Med 170, 256–261. [PMC free article] [PubMed] [Google Scholar]
- Malloy VL, Krajcik RA, Bailey SJ, Hristopoulos G, Plummer JD, and Orentreich N (2006). Methionine restriction decreases visceral fat mass and preserves insulin action in aging male Fischer 344 rats independent of energy restriction. Aging Cell 5, 305–314. [PubMed] [Google Scholar]
- Malloy VL, Perrone CE, Mattocks DA, Ables GP, Caliendo NS, Orentreich DS, and Orentreich N (2013). Methionine restriction prevents the progression of hepatic steatosis in leptin-deficient obese mice. Metabolism 62, 1651–1661. [PubMed] [Google Scholar]
- Matsui M, Oshima M, Oshima H, Takaku K, Maruyama T, Yodoi J, and Taketo MM (1996). Earlyembryonic lethality caused bytargeted disruption of the mouse thioredoxin gene. Dev. Biol 178, 179–185. [PubMed] [Google Scholar]
- Mickiewicz B, Villemaire ML, Sandercock LE, Jirik FR, and Vogel HJ (2014). Metabolic changes associated with selenium deficiency in mice. Biometals 27, 1137–1147. [PubMed] [Google Scholar]
- Miller RA, Buehner G, Chang Y, Harper JM, Sigler R, and Smith-Wheelock M (2005). Methionine-deficient diet extends mouse lifespan, slows immune and lens aging, alters glucose, T4, IGF-I and insulin levels, and increases hepatocyte MIF levels and stress resistance. Aging Cell 4, 119–125. [PMC free article] [PubMed] [Google Scholar]
- Misu H, Takayama H, Saito Y, Mita Y, Kikuchi A, Ishii KA, Chikamoto K, Kanamori T, Tajima N, Lan F, et al. (2017). Deficiency ofthe hepatokine selenoprotein P increases responsiveness to exercise in micethrough upregulation of reactive oxygen species and AMP-activated protein kinase in muscle. Nat. Med 23, 508–516. [PubMed] [Google Scholar]
- Mostert V, Hill KE, Ferris CD, and Burk RF (2003). Selective induction of liver parenchymal cell heme oxygenase-1 in selenium-deficient rats. Biol. Chem 384, 681–687. [PubMed] [Google Scholar]
- Müller M, Banning A, Brigelius-Flohé R, and Kipp A (2010). Nrf2 target genes are induced under marginal selenium-deficiency. Genes Nutr. 5, 297–307. [PMC free article] [PubMed] [Google Scholar]
- Nakayama A, Hill KE, Austin LM, Motley AK, and Burk RF (2007). All regions of mouse brain are dependent on selenoprotein P for maintenance of selenium. J. Nutr 137, 690–693. [PubMed] [Google Scholar]
- Neff F, Flores-Dominguez D, Ryan DP, Horsch M, Schröder S, Adler T, Afonso LC, Aguilar-Pimentel JA, Becker L, Garrett L, et al. (2013). Rapamycin extends murine lifespan but has limited effects on aging. J. Clin. Invest 123, 3272–3291. [PMC free article] [PubMed] [Google Scholar]
- Nonn L, Williams RR, Erickson RP, and Powis G (2003). The absence of mitochondrial thioredoxin 2 causes massive apoptosis, exencephaly, and early embryonic lethality in homozygous mice. Mol. Cell. Biol 23, 916–922. [PMC free article] [PubMed] [Google Scholar]
- Novoselov SV, Kim HY, Hua D, Lee BC, Astle CM, Harrison DE, Friguet B, Moustafa ME, Carlson BA, Hatfield DL, et al. (2010). Regulation of selenoproteins and methionine sulfoxide reductases A and B1 by age, calorie restriction, and dietary selenium in mice. Antioxid. Redox Signal 12, 829–838. [PMC free article] [PubMed] [Google Scholar]
- Ogawa-Wong AN, Berry MJ, and Seale LA (2016). Selenium and metabolic disorders: an emphasis on type 2 diabetes risk. Nutrients 8, 80. [PMC free article] [PubMed] [Google Scholar]
- Olson GE, Whitin JC, Hill KE, Winfrey VP, Motley AK, Austin LM, Deal J, Cohen HJ, and Burk RF (2010). Extracellular glutathione peroxidase (Gpx3) binds specifically to basement membranes of mouse renal cortex tubule cells. Am. J. Physiol. Renal Physiol 298, F1244–F1253. [PMC free article] [PubMed] [Google Scholar]
- Oster O, Schmiedel G, and Prellwitz W (1988). Theorgan distribution of selenium in German adults. Biol. Trace Elem. Res 15, 23–45. [PubMed] [Google Scholar]
- Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, Sethumadhavan S, Woo HK, Jang HG, Jha AK, et al. (2011). Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature 476, 346–350. [PMC free article] [PubMed] [Google Scholar]
- Prigge JR, Coppo L, Martin SS, Ogata F, Miller CG, Bruschwein MD, Orlicky DJ, Shearn CT, Kundert JA, Lytchier J, et al. (2017). Hepatocyte hyperproliferation upon liver-specific co-disruption of thioredoxin-1, thioredoxin reductase-1, and glutathione reductase. Cell Rep. 19, 2771–2781. [PMC free article] [PubMed] [Google Scholar]
- Raines AM, and Sunde RA (2011). Selenium toxicity but not deficient or super-nutritional selenium statusvastly altersthetranscriptome in rodents. BMC Genomics 12, 26. [PMC free article] [PubMed] [Google Scholar]
- Ran Q, Liang H, Ikeno Y, Qi W, Prolla TA, Roberts LJ 2nd, Wolf N, Van Remmen H, and Richardson A (2007). Reduction in glutathione peroxidase 4 increases life span through increased sensitivity to apoptosis. J. Gerontol. A Biol. Sci. Med. Sci 62, 932–942. [PubMed] [Google Scholar]
- Rayman MP (2012). Selenium and human health. Lancet 379, 1256–1268. [PubMed] [Google Scholar]
- Rayman M (2017). Effect of long-term selenium supplementation on mortality: results from a multiple-dose, randomised controlled trial. Free Radic. Biol. Med 127, 46–54. [PubMed] [Google Scholar]
- Rayman MP, and Stranges S (2013). Epidemiology of selenium and type 2 diabetes: can we make sense of it? Free Radic. Biol. Med 65, 1557–1564. [PubMed] [Google Scholar]
- Reid ME, Duffield-Ullico AJ, Slate E, Natarajan N, Turnbull B, Jacobs E, Combs GF Jr., Alberts DS, Clark LC, and Marshall JR (2008). The nutritional prevention of cancer: 400 mcg per day selenium treatment. Nutr. Cancer 60, 155–163. [PubMed] [Google Scholar]
- Richie JP Jr., Leutzinger Y, Parthasarathy S, Malloy V, Orentreich N, and Zimmerman JA (1994). Methionine restriction increases blood glutathione and longevity in F344 rats. FASEB J. 8, 1302–1307. [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47. [PMC free article] [PubMed] [Google Scholar]
- Schmidt EE (2015). Interplay between cytosolic disulfide reductase systems and the Nrf2/Keap1 pathway. Biochem. Soc. Trans 43, 632–638. [PMC free article] [PubMed] [Google Scholar]
- Scranton PE Jr., and McDermott JE (1997). Pathologic anatomic variations in subtalar anatomy. Foot Ankle Int. 18, 471–476. [PubMed] [Google Scholar]
- Seale LA, Gilman CL, Hashimoto AC, Ogawa-Wong AN, and Berry MJ (2015). Diet-induced obesity in theselenocysteine lyase knockout mouse. Antioxid. Redox Signal 23, 761–774. [PMC free article] [PubMed] [Google Scholar]
- Sinha I, Karagoz K, Fogle RL, Hollenbeak CS, Zea AH, Arga KY, Stanley AE, Hawkes WC, and Sinha R (2016). “Omics” of selenium biology: a prospective study of plasma proteome network before and after selenized-yeast supplementation in healthy men. OMICS 20, 202–213. [PubMed] [Google Scholar]
- Soerensen J, Jakupoglu C, Beck H, Förster H, Schmidt J, Schmahl W, Schweizer U, Conrad M, and Brielmeier M (2008). The role of thioredoxin reductases in brain development. PLoS ONE 3, e1813. [PMC free article] [PubMed] [Google Scholar]
- Stone KP, Wanders D, Orgeron M, Cortez CC, and Gettys TW (2014). Mechanisms of increased in vivo insulin sensitivity by dietary methionine restriction in mice. Diabetes 63, 3721–3733. [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, and Mesirov JP (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550. [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Kuehn H, Gould J, Tamayo P, and Mesirov JP (2007). GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics 23, 3251–3253. [PubMed] [Google Scholar]
- Sunde RA, Raines AM, Barnes KM, and Evenson JK (2009). Selenium status highly regulates selenoprotein mRNA levels for only a subset of the selenoproteins in the selenoproteome. Biosci. Rep 29, 329–338. [PMC free article] [PubMed] [Google Scholar]
- Takata Y, Kristal AR, Santella RM, King IB, Duggan DJ, Lampe JW, Rayman MP, Blount PL, Reid BJ, Vaughan TL, and Peters U (2012). Selenium, selenoenzymes, oxidative stress and risk of neoplastic progression from Barrett’s esophagus: results from biomarkers and genetic variants. PLoS ONE 7, e38612. [PMC free article] [PubMed] [Google Scholar]
- Tanaka N, Takahashi S, Zhang Y, Krausz KW, Smith PB, Patterson AD, and Gonzalez FJ (2015). Role of fibroblast growth factor21 in the early stage of NASH induced by methionine- and choline-deficient diet. Biochim. Biophys. Acta 1852, 1242–1252. [PMC free article] [PubMed] [Google Scholar]
- Tsuji PA, Carlson BA, Anderson CB, Seifried HE, Hatfield DL, and Howard MT (2015). Dietary Selenium Levels Affect Selenoprotein Expression and Support the Interferon-γ and IL-6 Immune Response Pathways in Mice. Nutrients 7, 6529–6549. [PMC free article] [PubMed] [Google Scholar]
- Turanov AA, Kehr S, Marino SM, Yoo MH, Carlson BA, Hatfield DL, and Gladyshev VN (2010). Mammalian thioredoxin reductase 1: roles in redox homoeostasis and characterization of cellular targets. Biochem. J 430, 285–293. [PMC free article] [PubMed] [Google Scholar]
- Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, and Müller F (2003). Genetics: influence of TOR kinase on lifespan in C. elegans. Nature 426, 620. [PubMed] [Google Scholar]
- Vinceti M, Dennert G, Crespi CM, Zwahlen M, Brinkman M, Zeegers MP, Horneber M, D’Amico R, and Del Giovane C (2014). Selenium for preventing cancer. Cochrane Database Syst. Rev (3), CD005195. [PMC free article] [PubMed] [Google Scholar]
- Wanders D, Burk DH, Cortez CC, Van NT, Stone KP, Baker M, Mendoza T, Mynatt RL, and Gettys TW (2015). UCP1 is an essential mediator of the effects of methionine restriction on energy balance but not insulin sensitivity. FASEB J. 29, 2603–2615. [PMC free article] [PubMed] [Google Scholar]
- Wu JJ, Liu J, Chen EB, Wang JJ, Cao L, Narayan N, Fergusson MM, Rovira II, Allen M, Springer DA, et al. (2013). Increased mammalian lifespan and a segmental and tissue-specific slowing of aging after genetic reduction of mTOR expression. Cell Rep. 4, 913–920. [PMC free article] [PubMed] [Google Scholar]
- Wu S, Mariotti M, Santesmasses D, Hill KE, Baclaocos J, Aparicio-Prat E, Li S, Mackrill J, Wu Y, Howard MT, et al. (2016). Human selenoprotein P and S variant mRNAs with different numbers of SECIS elements and inferences from mutant mice of the roles of multiple SECIS elements. Open Biol. 6, 160241. [PMC free article] [PubMed] [Google Scholar]
- Wu RT, Cao L, Mattson E, Witwer KW, Cao J, Zeng H, He X, Combs GF Jr., and Cheng WH (2017). Opposing impacts on healthspan and longevity by limiting dietary selenium in telomere dysfunctional mice. Aging Cell 16, 125–135. [PMC free article] [PubMed] [Google Scholar]
- Xia J, and Wishart DS (2010). MetPA: a web-based metabolomics tool for pathway analysis and visualization. Bioinformatics 26, 2342–2344. [PubMed] [Google Scholar]
- Yim SH, Tobe R, Turanov AA, and Carlson BA (2018). Radioactive 75Se Labeling and Detection of Selenoproteins. Methods Mol. Biol 1661, 177–192. [PubMed] [Google Scholar]