PNAS Plus
Epigenetic alterations in longevity regulators, reduced life span, and exacerbated aging-related pathology in old father offspring mice
Kan Xie
aMolecular and Cellular Cognition Lab, German Center for Neurodegenerative Diseases (DZNE), 53127 Bonn, Germany;
Devon P. Ryan
aMolecular and Cellular Cognition Lab, German Center for Neurodegenerative Diseases (DZNE), 53127 Bonn, Germany;
Brandon L. Pearson
aMolecular and Cellular Cognition Lab, German Center for Neurodegenerative Diseases (DZNE), 53127 Bonn, Germany;
Kristin S. Henzel
aMolecular and Cellular Cognition Lab, German Center for Neurodegenerative Diseases (DZNE), 53127 Bonn, Germany;
Frauke Neff
cInstitute of Pathology, Helmholtz Zentrum München, German Research Center for Environmental Health, 85764 Neuherberg, Germany;
dGerman Mouse Clinic, Institute of Experimental Genetics, Helmholtz Zentrum München, 85764 Neuherberg, Germany;
Ramon O. Vidal
eComputational Systems Biology Lab, German Center for Neurodegenerative Diseases (DZNE), 37077 Göttingen, Germany;
Magali Hennion
eComputational Systems Biology Lab, German Center for Neurodegenerative Diseases (DZNE), 37077 Göttingen, Germany;
Isabelle Lehmann
aMolecular and Cellular Cognition Lab, German Center for Neurodegenerative Diseases (DZNE), 53127 Bonn, Germany;
Melvin Schleif
bSelective Vulnerability of Neurodegenerative Diseases Lab, German Center for Neurodegenerative Diseases (DZNE), 53127 Bonn, Germany;
Susanne Schröder
aMolecular and Cellular Cognition Lab, German Center for Neurodegenerative Diseases (DZNE), 53127 Bonn, Germany;
Thure Adler
dGerman Mouse Clinic, Institute of Experimental Genetics, Helmholtz Zentrum München, 85764 Neuherberg, Germany;
fInstitute for Medical Microbiology, Immunology and Hygiene, Technische Universität München, 81675 Munich, Germany;
Birgit Rathkolb
dGerman Mouse Clinic, Institute of Experimental Genetics, Helmholtz Zentrum München, 85764 Neuherberg, Germany;
gChair of Molecular Animal Breeding and Biotechnology, Gene Center, Ludwig-Maximilians-Universität München, 81377 Munich, Germany;
Jan Rozman
dGerman Mouse Clinic, Institute of Experimental Genetics, Helmholtz Zentrum München, 85764 Neuherberg, Germany;
hGerman Center for Diabetes Research (DZD), 85764 München-Neuherberg, Germany;
Anna-Lena Schütz
eComputational Systems Biology Lab, German Center for Neurodegenerative Diseases (DZNE), 37077 Göttingen, Germany;
Cornelia Prehn
dGerman Mouse Clinic, Institute of Experimental Genetics, Helmholtz Zentrum München, 85764 Neuherberg, Germany;
Michel E. Mickael
eComputational Systems Biology Lab, German Center for Neurodegenerative Diseases (DZNE), 37077 Göttingen, Germany;
Marco Weiergräber
iResearch Group Experimental Neuropsychopharmacology, Federal Institute for Drugs and Medical Devices, 53175 Bonn, Germany;
Jerzy Adamski
dGerman Mouse Clinic, Institute of Experimental Genetics, Helmholtz Zentrum München, 85764 Neuherberg, Germany;
jChair of Experimental Genetics, Center of Life and Food Sciences Weihenstephan, Technische Universität München, 85350 Freising-Weihenstephan, Germany;
Dirk H. Busch
fInstitute for Medical Microbiology, Immunology and Hygiene, Technische Universität München, 81675 Munich, Germany;
Gerhard Ehninger
kDepartment of Internal Medicine I, University Hospital Carl Gustav Carus, Technical University Dresden, 01307 Dresden, Germany;
Anna Matynia
lJules Stein Eye Institute, University of California, Los Angeles, CA, 90095;
Walker S. Jackson
bSelective Vulnerability of Neurodegenerative Diseases Lab, German Center for Neurodegenerative Diseases (DZNE), 53127 Bonn, Germany;
Eckhard Wolf
gChair of Molecular Animal Breeding and Biotechnology, Gene Center, Ludwig-Maximilians-Universität München, 81377 Munich, Germany;
Helmut Fuchs
dGerman Mouse Clinic, Institute of Experimental Genetics, Helmholtz Zentrum München, 85764 Neuherberg, Germany;
Valerie Gailus-Durner
dGerman Mouse Clinic, Institute of Experimental Genetics, Helmholtz Zentrum München, 85764 Neuherberg, Germany;
Stefan Bonn
eComputational Systems Biology Lab, German Center for Neurodegenerative Diseases (DZNE), 37077 Göttingen, Germany;
mGerman Center for Neurodegenerative Diseases (DZNE), 72076 Tübingen, Germany;
nCenter for Molecular Neurobiology, University Medical Center Hamburg-Eppendorf, 20251 Hamburg, Germany
Martin Hrabě de Angelis
dGerman Mouse Clinic, Institute of Experimental Genetics, Helmholtz Zentrum München, 85764 Neuherberg, Germany;
hGerman Center for Diabetes Research (DZD), 85764 München-Neuherberg, Germany;
jChair of Experimental Genetics, Center of Life and Food Sciences Weihenstephan, Technische Universität München, 85350 Freising-Weihenstephan, Germany;
Dan Ehninger
aMolecular and Cellular Cognition Lab, German Center for Neurodegenerative Diseases (DZNE), 53127 Bonn, Germany;
Author contributions: M.W., J.A., D.H.B., G.E., W.S.J., E.W., H.F., V.G.-D., S.B., M.H.d.A., and D.E. designed research; K.X., D.P.R., B.L.P., K.S.H., F.N., R.O.V., M.H., I.L., M.S., S.S., T.A., B.R., J.R., A.-L.S., C.P., M.E.M., A.M., and D.E. performed research; K.X., D.P.R., B.L.P., K.S.H., F.N., R.O.V., M.H., M.S., T.A., B.R., J.R., C.P., M.E.M., A.M., and D.E. analyzed data; and D.P.R., B.L.P., S.B., and D.E. wrote the paper.
Associated Data
Significance
Aging-associated diseases are increasingly common in an aging global population. However, the contributors and origins of differential risk for unhealthy aging remain poorly understood. Using a mouse model, we found that offspring of aged fathers exhibited a reduced life span and more pronounced aging-associated pathologies than animals sired by young fathers. Tissue of offspring and aged fathers revealed shared epigenetic signatures and showed altered activation states of longevity-related cell signaling. Our results suggest that variability in aging trajectories could derive, in part, from the age at conception of the father, a possibility that warrants human epidemiological investigation.
Abstract
Advanced age is not only a major risk factor for a range of disorders within an aging individual but may also enhance susceptibility for disease in the next generation. In humans, advanced paternal age has been associated with increased risk for a number of diseases. Experiments in rodent models have provided initial evidence that paternal age can influence behavioral traits in offspring animals, but the overall scope and extent of paternal age effects on health and disease across the life span remain underexplored. Here, we report that old father offspring mice showed a reduced life span and an exacerbated development of aging traits compared with young father offspring mice. Genome-wide epigenetic analyses of sperm from aging males and old father offspring tissue identified differentially methylated promoters, enriched for genes involved in the regulation of evolutionarily conserved longevity pathways. Gene expression analyses, biochemical experiments, and functional studies revealed evidence for an overactive mTORC1 signaling pathway in old father offspring mice. Pharmacological mTOR inhibition during the course of normal aging ameliorated many of the aging traits that were exacerbated in old father offspring mice. These findings raise the possibility that inherited alterations in longevity pathways contribute to intergenerational effects of aging in old father offspring mice.
The concept of an immortal germ line posits that gametes persist through generations and, accordingly, are spared accumulated deterioration intrinsic to disposable somatic cell lineages (1, 2). This phenomenon is contingent on enhanced repair and rejuvenation in the immortal germ line (3) but nevertheless remains paradoxical considering evidence that sperm contribute genetic and epigenetic information to the zygote that can be incompletely erased (4) and is sensitive to parental experience (5) with oftentimes detrimental effects. In particular, epigenetic alterations in sperm are increasingly implicated in beneficial and deleterious influences in the offspring generation.
Advanced age is a risk factor for a range of disorders within an individual, including neurodegenerative diseases, cardiovascular diseases, and cancers but may also increase disease risk in the next generation. Epidemiological studies have shown an elevated risk for neuropsychiatric disorders, as well as other diseases, in the offspring of fathers with an advanced age at conception compared with the offspring of young fathers (6–8). In addition to possible mutagenic events, aging is associated with widespread epigenetic changes (9–14). Epigenetic alterations in the male germ line, induced by dietary and other environmental factors (5, 15–17), can elicit long-lasting effects on a broad range of physiological states in the next generation. Thus, alterations to paternal gametes could, in principle, influence aging and associated disease risks in the subsequent generation.
We set out to explore, in a mouse model, whether advanced paternal age influences life span and aging-associated phenotypes in the offspring generation. We combined genome-wide epigenetic analyses of sperm in the parental generation with epigenomic, transcriptomic, and biochemical studies as well as extensive phenotypic analyses in the offspring of old (>21 mo old) vs. young (4 mo old) fathers. We provide evidence that old father offspring (OFO) display a reduced life span and an exacerbation of aging-associated phenotypes accompanied by numerous epigenetic alterations in the paternal germ line as well as offspring tissue.
Results
Reduced Life Span and Exacerbated Aging-Associated Pathology in OFO Mice.
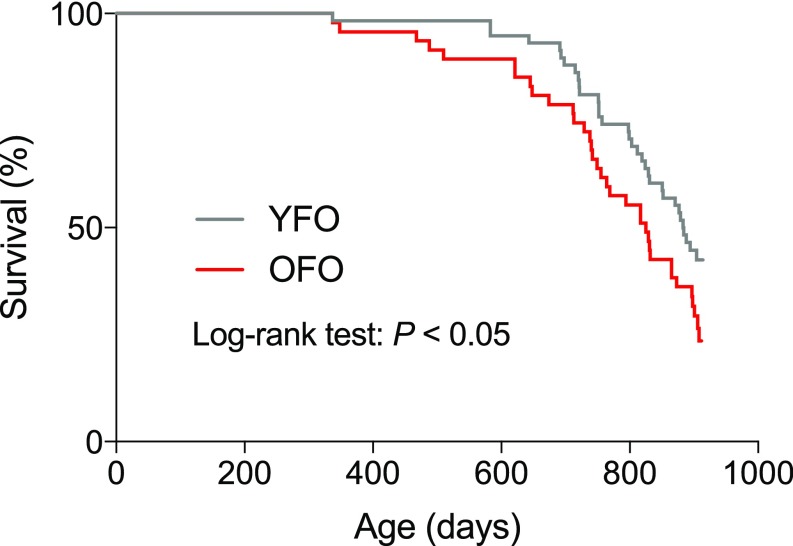
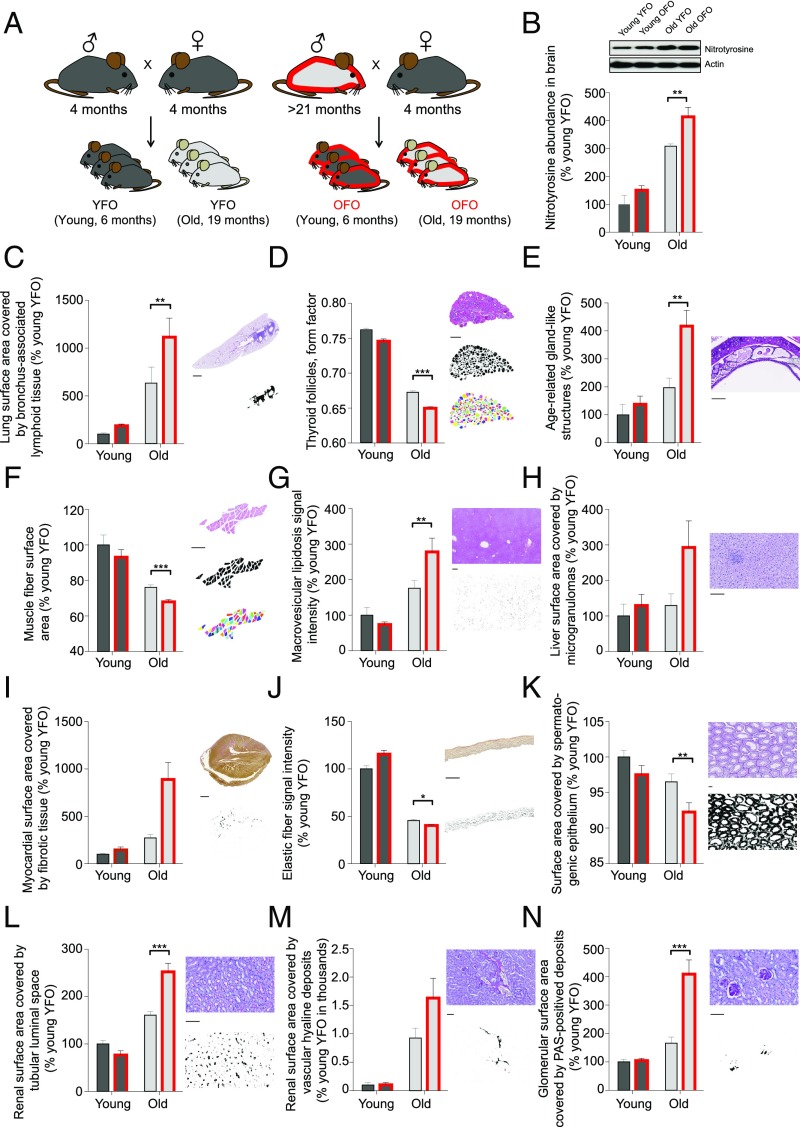
To examine whether advanced paternal age has effects on longevity, we measured natural life span in OFO mice as well as young father offspring (YFO) controls. Log-rank test-based analysis of life span data revealed a significant effect of paternal age (P < 0.05) with a ∼6.6% reduction of median life span in OFO mice relative to YFO animals (OFO: 825 d; YFO: 883.5 d) (Fig. 1; for additional analyses of life span data, see SI Appendix, Fig. S1). To address whether advanced paternal age affects aging-associated pathology, we analyzed a large cohort of aged offspring mice (19 mo old) of young and old fathers, along with the corresponding young adult reference group (6 mo old), and analyzed a number of histopathological and molecular aging traits across various tissues and organ systems. These studies included a quantitative assessment of aging traits in heart (myocardial fibrosis) (18), arteries (elastic fiber content of the arterial wall) (19), kidney (glomerulosclerosis, tubular atrophy, vascular hyalinosis) (20), skeletal muscle (muscle atrophy) (21), testis (testicular atrophy) (22), trachea (age-related retention cysts in the submucosa) (23), lung (bronchus-associated lymphoid tissue) (24), liver (macrovesicular lipidosis, microgranulomas) (25), thyroid gland (altered follicle morphology) (26), and brain (nitrotyrosine abundance) (27). Our studies revealed that many of the aging traits examined were more pronounced in OFO mice than in YFO animals (Fig. 2), indicating that advanced paternal age exacerbated specific aging-associated tissue changes. Together, our findings show that advanced paternal age in mice was associated with a reduced life span and an exacerbation of a range of tissue aging phenotypes.
OFO mice exhibited a reduced life span compared with offspring of young fathers. Survival curves were calculated for OFO (n = 47) and YFO mice (n = 58). Log-rank test showed a significant effect of paternal age on life span (P = 0.0465).
Exacerbation of aging traits in OFO. (A) Experimental design. (B) Nitrotyrosine abundance, as determined by Western blot, in brain homogenates of aged YFO and OFO animals (old YFO, n = 6 mice from six litters; old OFO, n = 6 mice from six litters) and young adult reference groups (young YFO, n = 6 mice from six litters; young OFO, n = 6 mice from six litters) (age: P < 0.0001; paternal age: P = 0.006; interaction: P = 0.30). (C–N) Histopathological aging traits examined in aged YFO and OFO animals (old YFO, n = 37 mice from 12 litters; old OFO, n = 45 mice from 17 litters) as well as young adult reference groups (young YFO, n = 16 mice from six litters; young OFO, n = 14 mice from seven litters). Shown are examples of histological images, probability maps (generated via automated segmentation of histological images), and if applicable, primary object identification maps, along with the quantification of the respective image features. (C) Bronchus-associated lymphoid tissue (age: P < 0.0001; paternal age: P = 0.02; interaction: P = 0.10); (D) thyroid follicle morphology (form factor; a value of 1 reflecting a perfectly circular structure; values between 1 and 0 report progressively more irregularly shaped structures) (age: P < 0.0001; paternal age: P < 0.0001; interaction: P = 0.18); (E) age-related gland-like structures in the tracheal submucosa (age: P = 0.0008; paternal age: P = 0.02; interaction: P = 0.10); (F) skeletal muscle fiber atrophy (age: P < 0.0001; paternal age: P = 0.007; interaction: P = 0.82); (G) macrovesicular lipidosis affecting the liver (age: P < 0.0001; paternal age: P = 0.14; interaction: P = 0.02); (H) liver microgranulomas (age: P = 0.18; paternal age: P = 0.18; interaction: P = 0.35); (I) myocardial fibrosis (age: P = 0.02; paternal age: P = 0.08; interaction: P = 0.13); (J) elastic fiber content of arterial walls (age: P < 0.0001; paternal age: P = 0.02; interaction: P < 0.0001); (K) testis atrophy (age: P = 0.004; paternal age: P = 0.03; interaction: P = 0.55); (L) renal tubular atrophy (age: P < 0.0001; paternal age: P = 0.046; interaction: P = 0.001); (M) renal vascular hyalinosis (age: P = 0.0003; paternal age: P = 0.27; interaction: P = 0.27); (N) glomerulosclerosis (age: P = 0.0002; paternal age: P = 0.01; interaction: P = 0.02). [Scale bars: 1 mm (C and I); 300 µm (D and G); 100 µm (E, F, and H–N).] *P < 0.05; **P < 0.01; ***P < 0.001. Graphs show mean ± SEM.
Epigenetic Analyses Identified Aging-Associated Changes in Longevity Regulators in Aged Sperm.
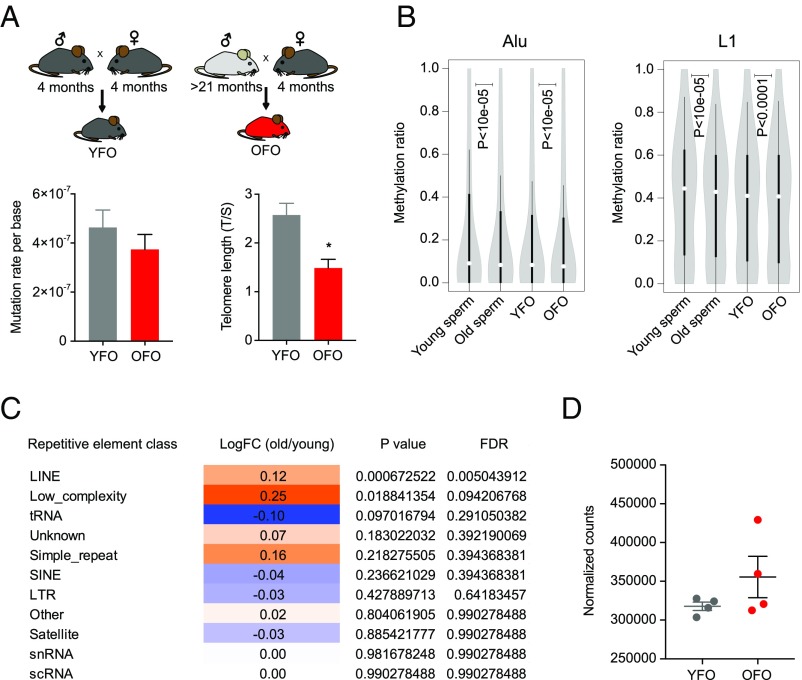
Intergenerational effects of advanced paternal age could involve an accumulation of mutations in aging germ cells that are subsequently inherited by the next generation. We estimated mutation rates in the offspring of young and old fathers by calling private variants in offspring RNA-sequencing (RNA-seq) data. These analyses yielded mutation rate estimates ranging from ∼1 to ∼7e-7 mutations per base (Fig. 3A). We were unable to discern group differences, suggesting that an age-related accumulation of point mutations in the male germ line may not account for the intergenerational effects of aging described here. It is also conceivable that aging-associated erosion of telomere length in old fathers could have been transferred to offspring, thus contributing to their premature aging phenotypes. Telomere length in the OFO hippocampus was indeed shorter than in YFO, supporting the notion that offspring phenotypes could be sensitive to inherited changes in telomere length (Fig. 3A).
Assessment of mutation, telomeres, and repetitive elements in paternal age effect. (A) To estimate mutation rates, we called private variants in RNA-seq data obtained from YFO and OFO (n = 6 mice from six litters each per condition). We found no determinable difference between the paternal age groups (P = 0.37). Telomere length was shorter in brain tissue of OFO mice (n = 3 mice per group). (B) DNA methylation was reduced in old father sperm and in OFO tissue. Violin plots show distributions of cytosine methylation ratios across Alu and LINE-1 elements in aged (24 mo old, pool of five mice) vs. young sperm (4 mo old, pool of five mice) and between OFO (pool of 10 mice) vs. YFO (pool of 10 mice) mice. (C) Table displaying results of RNA-seq–based repeat expression analysis in old (22 mo, n = 3 mice) vs. young (3 mo, n = 8 mice) sperm, demonstrating increased expression of LINE elements. (D) Analysis of LINE expression in OFO vs. YFO mice (n = 4 mice per group, 4 wk old, hippocampus) indicates a potential increase but without reaching statistical significance (P = 0.057). Graphs, unless otherwise stated, display mean ± SEM.
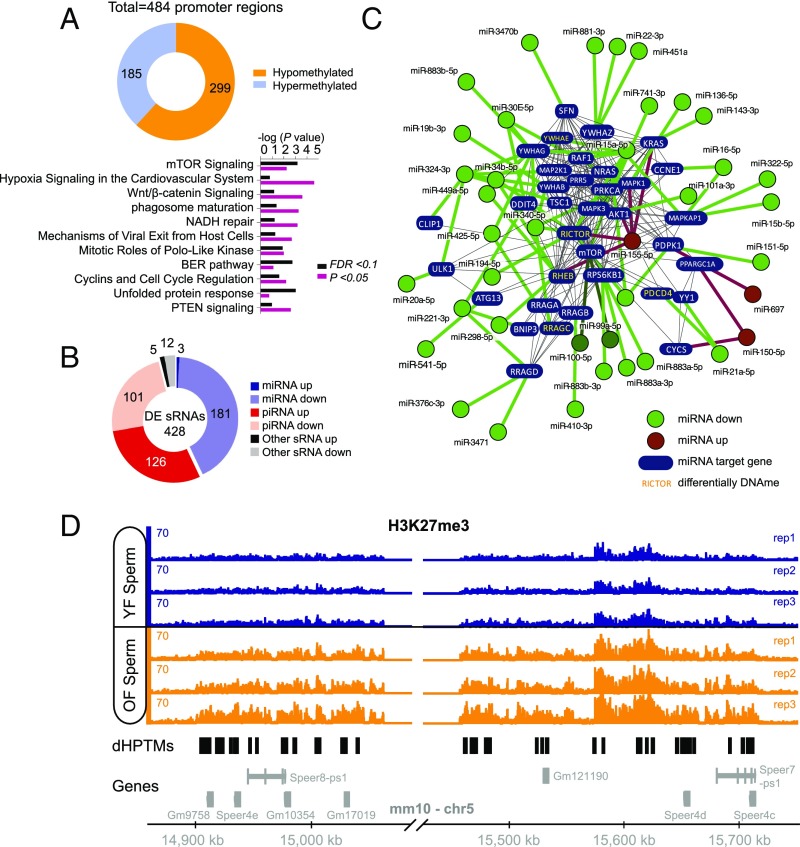
In addition to possible mutagenic events, aging is associated with widespread epigenetic changes (9–14). Because reprogramming of the epigenome during embryonic development is thought to be incomplete (4), it is conceivable that a subset of aging-associated epigenetic alterations survives reprogramming of the paternal epigenome and is, hence, passed on to the next generation. We employed reduced representation bisulfite sequencing (RRBS) to assess, in a genome-wide fashion and with single-base resolution, CpG-rich promoter-based DNA methylation in sperm of young and old males. These analyses revealed widespread promoter methylation changes (n = 484 promoter regions, FDR < 0.1; Fig. 4A and SI Appendix, Table S1), 299 of which were significantly hypomethylated (∼62%) and 185 significantly hypermethylated (∼38%) in sperm of old males. Pathway analysis identified a number of pathways with significant enrichment among promoters with differentially methylated regions in aged sperm, including several pathways with relevance to senescence, aging, and longevity, such as mTOR signaling, PTEN signaling, IGF1 signaling, p53 signaling, and immune-regulatory pathways (Fig. 4A and SI Appendix, Tables S2 and S3).
Epigenetic changes in aged sperm implicated alterations in the mTOR pathway. (A) Whole genome, single-base resolution methylome analyses (RRBS) identified differentially methylated promoters in aged sperm. Shown is the total number of differentially methylated regions (FDR < 0.1) within promoters and significantly enriched canonical pathways among the genes with differentially methylated (FDR < 0.1 or P < 0.05) regions in their promoters. (B) Differentially expressed sRNAs in sperm. (C) Interaction network of differentially expressed miRNAs targeting components on the mTOR pathway. Green denotes miRNAs down-regulated in aged sperm; red indicates miRNAs up-regulated in aged sperm. Genes denoted in yellow additionally contained a differentially methylated region within their promoter (which were all hypomethylated in aged sperm, except for Pdcd4, which contained a hypermethylated promoter region). (D) Identified hot spot for H3K27me3 modification differences between old and young sperm. This whole region on chromosome 5 featured an increased signal of H3K27me3 in old father samples compared with young father samples (black regions under horizontal axis indicate loci of differential histone posttranslational modifications, or dHPTMs). Most of the genes in this region belong to a family of spermatogenesis-related genes (Speer).
Aging-associated DNA methylation changes can influence promoter areas, but notable changes also occur within noncoding, repetitive regions of the genome, where DNA methylation is thought to play a role in transposon silencing (28). Our DNA methylation analyses showed an overall loss of methylation across Alu and LINE-1 elements in aged relative to young sperm (Fig. 3B), which is consistent with evidence of widespread aging-associated hypomethylation of these genomic regions (10, 29) and an increased expression of transposable elements during aging (10, 30).
Small noncoding RNA populations in sperm have also been implicated in the intergenerational modulation of phenotypes in offspring animals (15, 31). To identify possible age effects on the abundance of small RNAs in sperm, we performed small RNA-seq analyses of sperm from young and old animals. In total, 428 sRNAs were differentially expressed (adjusted P < 0.05), of which 184 (43%) were microRNAs (miRNAs) and 227 (53%) were piwi-interacting RNAs (piRNAs) (Fig. 4B and SI Appendix, Table S4). Of the 459 miRNAs that were detected in sperm, 184 (∼40%) were differentially expressed, 181 (∼98%) of which were down-regulated in sperm of old males. High confidence validated targets of miRNAs altered in aged sperm were significantly enriched for signaling pathways including mTOR signaling, insulin, and growth factor signaling (Fig. 4C and SI Appendix, Table S5). Interestingly, the down-regulation of miRNAs that target components of mTOR-related cell signaling, including the known mTOR regulators miR99, miR100, and miR15, is consistent with the hypomethylation within several promoters of mTOR-related genes, pointing to increased mTOR activity in the germ line of old males (Fig. 4C). Further analyses established an increased expression of LINE repeats in aged sperm (Fig. 3 B and C), consistent with the aging-associated hypomethylation of these genomic regions and changes in piRNA expression that we observed.
Next, we examined possible changes in chromatin composition and structure in aged sperm. Histone/protamine exchange is known to be incomplete during spermatogenesis, and consequently, histones are partially retained in sperm chromatin of mice and humans (32). If, and to what extent, histone retention in sperm may be altered during aging is unknown. We used ChIP-seq to identify possible aging-associated alterations in sperm histone occupancy, focusing on H3K4me3 and H3K27me3 retention (SI Appendix, Fig. S2). We identified a small region on chromosome 5 that contained 90% of the differential histone posttranslational modifications (Fig. 4D; SI Appendix, Table S6). This region also carried many differentially methylated regions in aged sperm and corresponded to a previously noted hotspot for differentially methylated regions in aging murine hematopoietic stem cells (33). Most of the genes in this region belong to a family of spermatogenesis-related genes (Speer). In conclusion, the analyses described above identified notable and consistent epigenetic changes in aged sperm enriched in pathways implicated in the regulation of senescence, aging, and longevity.
Epigenetic Analyses Uncovered Changes in Longevity Regulators in OFO.
Given the age-associated epigenetic changes in sperm described above, we next investigated if similar changes exist in DNA methylation in tissue of OFO mice. To identify possible DNA methylation changes in OFO animals (Fig. 5A), we performed RRBS on tissue harvested from YFO and OFO mice (hippocampus at 4 wk of age). These analyses revealed a set of promoters with differentially methylated regions in offspring tissue (n = 222 promoter regions, FDR < 0.1; Fig. 5B and SI Appendix, Table S7), 189 of which were significantly hypomethylated (∼85%) and 33 significantly hypermethylated (∼15%) in OFO animals. We noted 14 promoter regions that were also differentially methylated in aged sperm, which is more than expected by chance (P = 0.016). Pathway analyses showed enrichment for genes encoding components of mTOR-related cell signaling (i.e., mTOR signaling, eIF4E and p70S6K signaling) as well as of immune-regulatory pathways among the differentially methylated promoters in OFO (Fig. 5C and SI Appendix, Tables S3 and S8). Akin to the findings in aged sperm, we also observed an overall reduction of Alu and LINE-1 methylation in OFO animals compared with YFO controls and a possible increase of LINE expression in OFO animals (Fig. 3D). Interestingly, a prior study in 129SvEv/Tac mice showed overall loss of DNA methylation at TSS flanking regions in the offspring of fathers of an intermediate age range (12–14 mo) (9). Together, these studies identified similarities between epigenetic changes in aged sperm and OFO tissue.
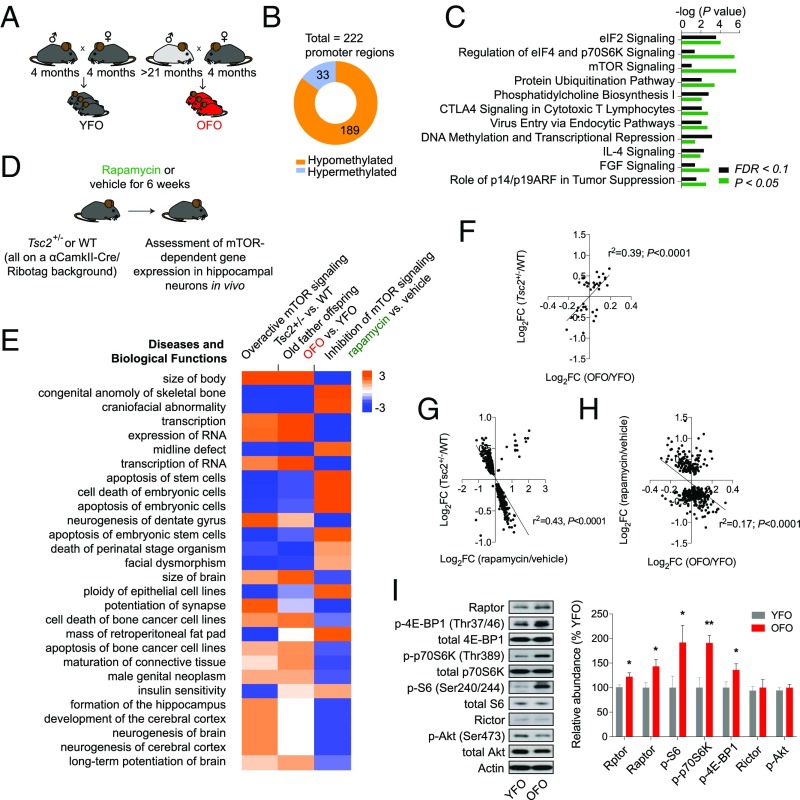
Overactive mTOR signaling in OFO. (A) Experimental design; analyses of genome-wide DNA methylation changes, transcriptional effects, and signaling alterations were carried out in tissue derived from 4-wk-old OFO and YFO mice. (B) Whole-genome, single-base resolution methylome analyses (RRBS) identified differentially methylated promoters in F1 offspring tissue (YFO vs. OFO). Shown is the total number of differentially methylated (FDR < 0.1) regions within promoters, proportions of hypo- vs. hypermethylated regions, as well as (C) significantly enriched canonical pathways among the genes with differentially methylated (FDR < 0.1 or P < 0.05) regions in their promoters. (D) We used the Ribotag technology together with RNA-seq to define hippocampal gene expression changes associated with activation (Tsc2+/− mutation) and inhibition (rapamycin) of the mTOR signaling pathway. (E) Enriched pathways from RNA-seq experiments revealed correspondence in Tsc2+/− mutants and OFO animals, with largely inverted changes in rapamycin-treated animals: Gene expression changes induced by the Tsc2+/− mutation and rapamycin predicted oppositional effects on biological functions, such as body size and transcriptional regulation; predicted effects of OFO-related gene expression changes resembled those associated with the Tsc2+/− genotype. Positive z scores, shown in the table, indicate activating effects, while negative z scores imply inhibitory action on the corresponding biological process. (F) Significant, positive correlation between gene expression in mTOR-hyperactive Tsc2+/− mutants (FDR < 0.1) and OFO mice vs. their respective controls. (G) Gene expression changes (P < 0.05) induced by rapamycin or Tsc2 haploinsufficiency showed an inverse correlation. (H) Transcriptional influences of rapamycin (FDR < 0.1) were inversely correlated with OFO mice vs. their respective controls. (I) Western blot experiments revealed an increased activation state of mTORC1 signaling along with an elevated expression of Rptor/Raptor in OFO hippocampus (p-p70S6K, p-4E-BP1, p-S6: n = 12 mice from 12 litters per group; Raptor: n = 6 mice from six litters per group). The bar graph shows the phosphorylation status (i.e., phosphoprotein normalized to total protein) of the mTORC1 downstream effectors ribosomal protein S6, p70S6K, and 4E-BP1, as well as the expression of Rptor (mRNA level analysis) and Raptor (protein level analysis). Also shown are the expression levels of Rictor (n = 6 mice from six litters per group) and the phosphorylation status of Akt at the mTORC2-sensitive site Ser473 (n = 12 mice from 12 litters per group). Graphs show mean ± SEM. *P < 0.05; **P < 0.01.
Next, we asked whether the epigenetic changes described above were associated with alterations in gene expression in OFO mice. RNA-seq–based transcriptome analyses of hippocampal tissue from 4-wk-old offspring of young and old fathers revealed a number of differentially expressed (FDR < 0.1) genes, some of which play roles in the regulation of immune processes (Ccl24, Ccl2, Il13ra2) (34–36) or neuropsychiatric disease risk (Gpr88) (37) (SI Appendix, Table S9). Broader pathway analysis of 720 genes (P < 0.05) predicted an activation of immune and inflammatory processes in OFO tissue (e.g., downstream targets of Ifn-γ and lipopolysaccharide were altered in a manner consistent with an activation of these immune regulators and downstream effector processes) and also highlighted a significant enrichment of additional parameters, such as cell growth and proliferation, cancer, and organismal survival (SI Appendix, Fig. S3). Notably, this is akin to transcriptional alterations observed in aging tissues that also feature changes in processes related to immunity, inflammation, cell growth, and proliferation (SI Appendix, Fig. S4 and Table S10) (38).
Overactive mTORC1 Signaling in OFO Mice.
The epigenetic analyses described above provide evidence for an alteration of specific pathways and functions in aged sperm, as well as OFO mice (Figs. 3–5), including mTOR signaling, a major regulator of growth, proliferation, and the immune system (39). To identify possible novel downstream effectors of mTOR, translationally or transcriptionally regulated by mTOR-related cell signaling, we employed the Ribotag technology (SI Appendix, SI Methods) and RNA-seq to define and quantify mRNAs sensitive to modulation of mTOR signaling [using rapamycin to pharmacologically inhibit mTOR and a Tsc2+/− mutation to genetically overactivate mTOR signaling (40)] in hippocampal neurons in vivo (Fig. 5D). These analyses revealed a number of transcripts regulated by rapamycin and/or the Tsc2+/− mutation (Fig. 5E and SI Appendix, Tables S11 and S12): We identified n = 44 genes differentially expressed in the heterozygous Tsc2 mutants and n = 495 genes differentially expressed as a consequence of rapamycin treatment (FDR < 0.1). Gene expression changes in OFO hippocampus were positively correlated with those induced by the Tsc2+/− mutation and negatively correlated with those caused by rapamycin (Fig. 5 E–H), suggesting a gene expression signature in OFO mice that is associated with overactive mTOR signaling. To further examine if mTOR signaling is indeed altered in OFO, we performed Western blot analyses to quantify the phosphorylation status of ribosomal protein S6, the target of p70S6K downstream of mTORC1, at serine residues 240/244. These analyses revealed increased S6 phosphorylation in hippocampal tissue from OFO (Fig. 5I). Further studies also showed an increased phosphorylation of p70S6K (at Thr389) and 4E-BP1 (Thr37/46), two additional downstream effectors of mTORC1, as well as increased Raptor expression, a critical component of the rapamycin-sensitive mTORC1 protein complex, in OFO (Fig. 5I). In contrast, the expression of Rictor and the phosphorylation status of Akt at Ser473 were unaltered in OFO (Fig. 5I), indicating that mTORC2 signaling was not changed in OFO mice. To address whether the mTORC1 signaling changes were restricted to hippocampal tissue or were also evident in peripheral tissues, we assessed the phosphorylation status of the mTORC1 downstream effectors mentioned above (i.e., p70S6K, S6, 4E-BP1) in the periphery: Western blot-based analyses of lung and liver samples provided further evidence for increased mTORC1 signaling in OFO (SI Appendix, Fig. S5). Together, these results indicated that mTORC1 signaling was overactive in OFO mice.
We also addressed whether mTORC1 signaling is altered during aging in the male germ line. Western blot and immunocytochemical analyses did not reveal evidence for quantifiable levels of p-S6 (Ser240/244) in mature murine sperm. We next asked whether mTORC1 signaling is altered within spermatogenic precursor cells of aged animals compared with those of young adult controls. To address this question, we performed p-S6 (Ser240/244) immunofluorescence staining of testicular sections (SI Appendix, Fig. S6) and quantified the p-S6 signal normalized to the number of cells within seminiferous tubules of old versus young mice. Our studies showed an increased relative p-S6 (Ser240/244) signal in aged seminiferous tubules (SI Appendix, Fig. S6). The data provide evidence for an overactivation of the mTORC1 signaling pathway within the spermatogenic compartment of aged mice.
The Pharmacological mTOR Inhibitor Rapamycin Ameliorated Age-Related Transcriptional Changes and the Aging-Associated Pathologies Exacerbated in OFO Mice.
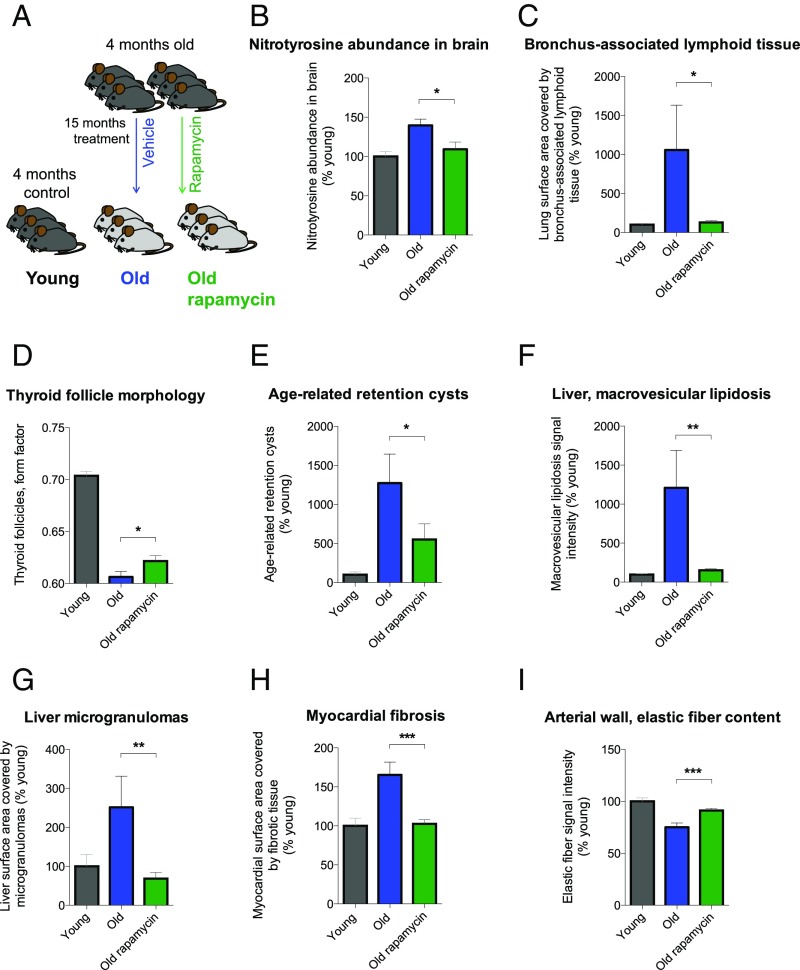
Next, we wanted to address whether the alterations in mTORC1 signaling in OFO mice could contribute to the exacerbated development of aging traits and the reduced life span in these animals (Figs. 1 and and2).2). mTOR is a well-established player in life span regulation (27, 41, 42) and has been shown to attenuate some mammalian aging traits by altering their rate of development and/or via symptomatic effects (27, 42). Moreover, chronic low-grade inflammatory processes (e.g., as a consequence of disinhibited mTORC1 signaling) may contribute to the development of aging-associated pathology (“inflammaging”) (43). We, therefore, set out to test whether aging traits altered in OFO are sensitive to treatment with the mTOR inhibitor rapamycin. RNA-seq–based studies performed on hippocampal tissue of aged mice (24 mo) treated with vehicle or rapamycin vs. young controls (4 mo) showed that rapamycin (for details, see Materials and Methods) effectively counteracted aging-associated transcriptional alterations in immune and inflammatory regulators (SI Appendix, Fig. S7 and Tables S13 and S14). To determine whether rapamycin prevents aging traits shown to be exacerbated in OFO mice, we treated 4-mo-old animals with rapamycin or vehicle control for a period of ∼15 mo (Fig. 6A). Detailed histopathological and molecular analyses revealed that many of the aging traits found to be exacerbated in OFO were ameliorated by rapamycin (Fig. 6), indicating that at least a subset of the aging traits that were exacerbated in old father offspring mice were sensitive to modulation of mTOR signaling.
Rapamycin ameliorated aging traits that were exacerbated in OFO. (A) Experimental design; (B) nitrotyrosine abundance in brain (n = 6 mice per group); (C) bronchus-associated lymphoid tissue; (D) thyroid follicle morphology; (E) age-related gland-like structures in the tracheal submucosa; (F) macrovesicular lipidosis affecting the liver; (G) liver microgranulomas; (H) myocardial fibrosis; (I) elastic fiber content of arterial walls. (C–I) Young, n = 12 mice; old, n = 10 mice; old rapamycin, n = 15 mice. Graphs show mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001.
To address whether rapamycin treatment of old fathers influences DNA methylation changes in their sperm, we subjected young (3 mo old) and old (22 mo old) mice to a 7-wk treatment with either rapamycin or vehicle control before sperm collection (SI Appendix, SI Results and Fig. S8). Targeted bisulfite sequencing of a ∼2-kb genomic region upstream of and overlapping with Gm7120 showed the expected age-related hypermethylation but no obvious treatment effect. It remains to be determined whether a longer term rapamycin treatment during the course of natural aging is sufficient to prevent age-dependent DNA methylation changes in sperm.
Metabolic, Immunological, and Behavioral Analyses of OFO Mice.
FACS profiling of peripheral blood leukocytes showed an increased abundance of several T cell subpopulations in OFO mice, including CD44high-expressing CD4+ T cells and CD44high-expressing CD8+ T cells (SI Appendix, SI Results and Fig. S9). Metabolic analyses indicated changes in the temporal plasma glucose profile following bolus glucose injection as well as elevated plasma cholesterol concentrations in OFO mice (SI Appendix, SI Results and Fig. S10). Behavioral analyses showed altered learning and memory in OFO animals (SI Appendix, SI Results and Fig. S11). These findings indicate that metabolic and immunological changes could contribute to altered aging trajectories in OFO mice.
Discussion
Here, we examined life span and aging-associated pathological changes in the F1 offspring of old vs. young male C57BL/6J Rj mice. We demonstrated that advanced paternal age was associated with a reduced life span and exacerbated molecular and histopathological aging traits in the next generation, indicating that paternal age exerted intergenerational effects on aspects of aging and age-related pathologies. These aging-associated alterations in OFO mice were associated with epigenetic, transcriptomic, and functional changes in life span-regulating pathways, including the mTORC1 pathway and immune regulators. Chronic treatment with the mTOR inhibitor and immunosuppressant rapamycin ameliorated a subset of the aging-associated phenotypes exacerbated in OFO mice, indicating that these aging traits were mTOR-sensitive and suggesting that the exacerbation of these phenotypes in OFO mice could be related to overactive mTORC1 signaling.
mTOR signaling is a key regulator of life span (27, 41, 42). Moreover, the activation states of mTOR-related cell signaling influence various aspects of aging and age-related pathologies, such as cancers (42, 44), cardiovascular disease (45), as well as pathological features in models of neurodegenerative disorders (46). mTOR’s influence on aging and age-related pathologies is likely mediated via broad effects on cell growth, proliferation, metabolism, immune functions, and proteostasis (46, 47). Future work needs to disentangle how specific cell biological consequences of mTOR inhibition are linked to individual aging-associated health outcomes.
mTOR inhibition-related life span extension has been observed in both male and female mice (27, 48). More pronounced life span effects in females fed a rapamycin-containing diet than in males may be due to sex effects on pharmacokinetic properties or food intake (resulting in increased blood concentrations in females at any given rapamycin dose) (48) rather than sex-specific roles of mTOR in life span regulation. While the analyses of our life span data did not provide support for an interaction between paternal age and sex, more work with larger animal numbers is needed to address whether effect sizes in males may differ from those in females.
In line with our current data, a previous report provided suggestive evidence, in a genetically heterogeneous stock of mice, of shorter lifespans in C57BL/6JIco-CBA/JIco hybrid mice sired by 120-wk-old fathers than in offspring conceived by younger fathers (49). Available data also suggest possible reductions in life span in Drosophila melanogaster (50) and in human offspring (51) sired by aged fathers. Studies using isogenic mice (as employed in the present study) are beneficial in their ability to exclude possible effects of paternal contact on offspring and the ability to limit genetic influences, as well as selection processes that can occur in experiments employing genetically heterogeneous populations.
At present, published evidence for enhanced risk for aging-related disease and phenotypes in OFO is limited; however, suggestive data that parental age might influence offspring aging trajectories exist. Available evidence in human populations indicates that advanced paternal age may be associated with adverse lipid profiles (increased LDL-cholesterol and total cholesterol/HDL-cholesterol ratio) (52) and, possibly, an elevated risk for obesity in young adulthood (53). Likewise, in the present mouse study, we also observed alterations in metabolic features (altered temporal profile of plasma glucose concentration in glucose tolerance test, plasma cholesterol concentration) in OFO mice. Whether these OFO mouse phenotypes are linked to metabolic changes related to altered mTOR signaling (39) needs to be addressed in future studies. Increased mTORC1 activity could, via SREBP2 activation, be associated with elevated cholesterol biosynthesis, as well as increased intestinal cholesterol uptake, which might contribute to elevated plasma cholesterol concentrations (although enhanced LDL receptor-dependent cellular uptake may have a lowering effect on plasma cholesterol) (54, 55). Further research is also necessary to determine if differential risk for cardiovascular and metabolic traits in humans (or for other age-related phenotypes/pathologies) derives from epigenetic factors such as those identified here in mice.
Paternal age at conception has increased over modern history (56). In the context of the present study, we examined the influence of fathers of a very advanced age (>21 mo old). It is currently unclear whether fathers of intermediate ages (e.g., 12 mo old), more typical of much of the murine advanced paternal age literature, cause exacerbated aging traits in their offspring. Similarly, future studies should assess more closely the possible advanced paternal age effects on specific aging-related pathologies in humans and examine the temporal dynamics and origins of age-related epigenetic changes within the paternal germ line.
Advanced paternal age effects could in general derive from a number of factors pre- and postconception. A prevalent hypothesis is that an age-related accumulation of mutations in the paternal germ line confers liability to offspring of older fathers (57). We found no evidence of higher mutation rates in tissue derived from OFO mice in our experimental context. However, since we estimated the mutation rate from RNA-seq data, which are limited to transcribed sequences, our data should be interpreted with caution given that mutations outside of coding regions could influence offspring phenotypes. Similarly, aging-associated erosion of telomeres has been cited as a potential central mediator of cellular aging (58). Human telomere lengths in somatic cells have a tendency to decrease with age, while sperm telomeres increase with age (59). Interestingly, offspring of older fathers in humans may have longer telomeres in their white blood cells and in sperm than offspring of younger fathers (59, 60), although it was suggested that this observation could have been due to a birth-cohort effect (61). Comparable data are limited in mice, but published evidence indicates that somatic and sperm telomeres decrease in aged mice (12 mo old) relative to young controls, and offspring of those aged fathers also showed reduced telomere length in somatic and germ cell lineages (62). Our data, in offspring brain, support this previous study and endorse a model wherein offspring of older fathers inherit eroded telomeres. However, age-related telomere attrition may have generally restricted roles with regards to many features of natural organismal aging, is known to be highly variable between and within species, and may have limited impact in many inbred mouse strains given the relatively long telomeres compared with other species (63, 64).
Old fathers could also influence offspring generations through alterations in experience stemming from paternal quality (65). We attempted to limit this influence by preventing any contact of offspring with their fathers. However, dams could have influenced offspring development based on their evaluation of mate quality (65). Future research could eliminate this possibility by performing in vitro fertilization and embryo transfer to pseudopregnant dams using the sperm of old and young fathers.
Consistent with the epigenetic data and a recent transcriptome analysis performed on frontal cortical tissue of OFO mice (66), our RNA-seq studies revealed an altered expression of immune and inflammatory regulators in OFO mice. Our FACS analyses revealed an increased proportion of CD44high-expressing activated/memory T cells in OFO mice. CD44high-expressing T cells are inversely correlated with life span in mice (67), conforming to the general notion that chronic low-grade inflammatory processes contribute to aging and age-related pathology (43). The transcriptional and cellular changes in OFO mice are akin to similar findings in animals with altered mTOR signaling (42, 68), suggesting that aberrant mTOR signaling in OFO mice could contribute to these immune phenotypes.
Our epigenetic analyses identified sets of overlapping promoter methylation changes in aged sperm and OFO tissue. Interestingly, differentially methylated promoters were enriched for pathways implicated in life span regulation, including mTOR-related cell signaling. Chronic inflammatory changes (69, 70) and/or altered redox states (71), associated with mitochondrial dysfunction (72), could contribute to mTOR signaling abnormalities in aging and senescence. Our epigenetic analyses also implicated changes in immune system regulators in aged mice and OFO animals. Available data support the notion of a two-way interaction between aging-related DNA methylation changes and inflammation: On the one hand, inflammation can drive DNA methylation changes associated with advanced age (12, 13), and on the other hand, aging-associated DNA methylation changes contribute to altered immunological states associated with advanced age (12, 73, 74). The data are consistent with the notion that immune functions as well as the activation states of mTOR-related cell signaling and the corresponding downstream effectors are regulated epigenetically in the context of aging and age-related disease (12, 13, 73–76).
In sum, our study reveals intergenerational effects of advanced paternal age on conserved longevity regulatory pathways as well as on aging-associated phenotypes and life span in mice. These data provide a foundation for future high-resolution bisulfite sequencing analyses to define aging-associated epigenetic changes in human sperm and indicate that further studies are warranted to ascertain whether advanced paternal age increases the risk for aging-associated disorders in humans.
Materials and Methods
Mice.
To generate YFO and OFO, we mated young adult (∼4 mo old) and old (>21 mo old) C57BL/6J Rj male, respectively, with young adult (∼4 mo old) 129S6/SvEv female mice. We chose to perform experiments in F1 offspring animals on a C57BL/6J Rj × 129S6/SvEv F1 background because these mice are isogenic, yet they are expected to exhibit hybrid vigor associated with outbreeding (77). Males had contact with females only during mating and never had contact with their offspring. Offspring mice were left undisturbed (except for weekly cage change) with their dam and were weaned at 21 d of age. All mice were housed under specific pathogen-free conditions within individually ventilated cages in groups of two to four mice per cage. We kept animals on a 12:12 h light/dark cycle. Mice received water and food ad libitum. If not stated otherwise, male and female mice were used at a balanced ratio in the studies described here. In all cases, the experimenter was blinded to the animal group assignments.
Rapamycin Treatment.
Rapamycin treatment was analogous to a previously published protocol (41). In brief, rapamycin was purchased from LC Laboratories and microencapsulated by Southwest Research Institute using a spinning disk atomization coating process with Eudragit S100 (Röhm Pharma) as the coating material. TestDiet provided Purina 5LG6 food containing encapsulated rapamycin or just the coating material (vehicle control). The encapsulated rapamycin was administered at 14 mg/kg food. To test whether rapamycin ameliorated aging traits that were exacerbated in OFO, we treated male C57BL/6J Rj mice with either rapamycin or vehicle control diet, starting at 4 mo of age, for 15 mo before performing histopathological and molecular analyses (rapamycin-treated, n = 15 mice; vehicle control, n = 10 mice). Animals were analyzed side-by-side with a group of young adult (i.e., 6 mo old at the time of assessment) male C57BL/6J Rj mice kept on a vehicle control diet (n = 12 mice). For RNA-seq–based gene expression analyses, aged (n = 2 mice; ∼24 mo old) and young (n = 2 mice; ∼4 mo old) mice were treated with rapamycin or vehicle control for a period of 6 wk before sacrifice.
Histology.
Histopathological analyses of aging traits were carried out on a cohort of aged YFO and OFO animals (∼19 mo old; old YFO, n = 37 mice from 12 litters; old OFO, n = 45 mice from 17 litters) as well as young adult reference groups (∼6 mo old; young YFO, n = 16 mice from six litters; young OFO, n = 14 mice from seven litters). Mice were killed with CO2. Following dissection, organs were fixed in 4% buffered formalin and embedded in paraffin for histological examination. Four μm-thick sections of heart, aorta, muscle, lung, trachea, thyroid gland, liver, kidney, and testis were cut and stained with hematoxylin and eosin (H&E), Periodic acid Schiff stain (PAS), and/or Elastica van Gieson (EvG, Weigert’s stain). Slides were digitalized using a virtual slide system (NanozoomerHT2.0, Hamamatsu). Computer-assisted analyses [using the automated image segmentation software ilastik (78) and CellProfiler (79)] were carried out to measure quantitatively specific aspects of age-related tissue changes, as specified in the main text and the figures. Histopathological data from young/old YFO/OFO offspring were analyzed by two-way ANOVA with the between-subject factors age (young vs. old) and paternal age (YFO vs. OFO), followed by Fisher’s LSD test to compare the old YFO and old OFO group where appropriate. Histopathological data from young, old/vehicle, and old/rapamycin mice were analyzed by one-way ANOVA, followed by planned comparison (Sidak’s test) of the old/vehicle and old/rapamycin group if appropriate.
Hippocampus Extraction.
Following cervical dislocation, brains were removed and placed in PBS. Hemibrains were then separated and hippocampi then removed after removing the midbrain. Care was taken to ensure that the hippocampi were free of choroid plexus contamination. Following extraction, hippocampi were immediately flash-frozen in liquid nitrogen.
Isolation of Sperm and Testes.
After sacrifice, the lower abdomen was exposed, and the testes and epididymides were removed and placed in PBS. The fat pads affixed to the testes were removed, and the testes were then separated from the epididymides and flash-frozen in liquid nitrogen. The epididymides were then placed in a separate dish with PBS, sliced with scissors, and placed into PBS-filled Eppendorf tubes. Sperm were allowed 30 min at room temperature to swim out. After that time, the supernatant was removed and spun down in a separate tube to concentrate the sperm. After removal of excess PBS, sperm were then flash-frozen in liquid nitrogen. For sperm counts, a 20-µL aliquot of swim-out sperm was collected before pelleting and diluted 1:10 in PBS and counted in a Neubauer chamber and calculated as an average of two replicate measurements from a single epididymis.
Western Blots.
Hippocampal tissue, lung, and liver of F1 offspring derived from old and young fathers were homogenized in lysis buffer containing 50 mM Tris (pH 7.4), 150 mM NaCl, 1% Triton X-100 (Sigma Aldrich), 1× protease inhibitors (Roche), and 1× phosphatase inhibitors (Roche) and incubated on ice for 30 min. Homogenates were centrifuged at 10,000 g for 10 min at 4 °C, and the supernatant was used for further analysis. Protein concentration was determined by Pierce BCA protein assay kit (Thermo Fisher Scientific). If not described otherwise, 20 µg protein was loaded on 10% or 15% self-cast Tris–glycine gels for electrophoresis and transferred onto 0.2 µm polyvinylidene fluoride (PVDF) membrane. Following blocking with 1× PBS (Thermo Fisher Scientific) with 10% (wt/vol) low-fat milk powder (Carl Roth) for 1 h at room temperature, membranes were probed with the respective primary antibodies at 4 °C overnight. After multiple washing steps in PBS with 0.1% Tween-20 (Sigma Aldrich), horseradish peroxidase-conjugated secondary antibodies (goat anti-mouse IgG, 1:5,000; Dako; goat anti-rabbit IgG, 1:3,000; Promega) were applied for 1.5 h at room temperature. Membranes were washed again, and immunosignals were detected using enhanced chemiluminescence (Amersham ECL Western Blotting Detection Reagents; GE Healthcare). ImageJ software (version 1.44; US National Institute of Health) was used for densitometric analyses. Raptor and Rictor were normalized to the actin band of the same lane. Phosphorylated proteins [p-S6 (Ser240/244), p-p70S6K (Thr389), p-4E-BP1 (Thr37/46), p-Akt (Ser473)] were normalized to the respective total protein band on the same lane. Unpaired, two-tailed t tests were used to compare data across groups. Information regarding primary antibodies is provided in SI Appendix, SI Methods.
Statistics.
Statistical tests were performed using GraphPad Prism (v7) or R (v3.1.2), if not described otherwise. Unpaired t tests, one-way, and two-way ANOVAs were utilized as described.
Study Approval.
Mouse studies were approved by Landesamt für Natur, Umwelt und Verbraucherschutz Nordrhein-Westfalen; Regierung von Oberbayern (in accordance with the German Animal Health and Welfare Act, German Federal Law, §8 Abs. 1 TierSchG); or the Chancellor’s Animal Research Committee at the University of California, Los Angeles.
For full information regarding methods, see also SI Appendix, SI Methods.
Acknowledgments
The German Center for Neurodegenerative Diseases (DZNE) animal caretaker team, DZNE animal facility management, the German Mouse Clinic (GMC) technicians, as well as the GMC animal caretaker team provided expert technical assistance and valuable support. The study was supported by DZNE, the German Federal Ministry of Education and Research (Infrafrontier Grant 01KX1012), Helmholtz Zukunftsthema Aging and Metabolic Programming (AMPro), the German Center for Diabetes Research (DZD), and the Helmholtz Alliance for Mental Health in Ageing Society (HA-215).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Datasets have been deposited to the European Nucleotide Archive (ENA) (accession no. PRJEB23859).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1707337115/-/DCSupplemental.