PNAS Plus
Effects of rapamycin on growth hormone receptor knockout mice
Yimin Fang
aDepartment of Internal Medicine, Southern Illinois University School of Medicine, Springfield, IL, 62702;
Cristal M. Hill
aDepartment of Internal Medicine, Southern Illinois University School of Medicine, Springfield, IL, 62702;
Justin Darcy
aDepartment of Internal Medicine, Southern Illinois University School of Medicine, Springfield, IL, 62702;
Adriana Reyes-Ordoñez
bDepartment of Cell and Developmental Biology, University of Illinois Urbana–Champaign, Urbana, IL, 61801;
Edwin Arauz
bDepartment of Cell and Developmental Biology, University of Illinois Urbana–Champaign, Urbana, IL, 61801;
Samuel McFadden
aDepartment of Internal Medicine, Southern Illinois University School of Medicine, Springfield, IL, 62702;
Chi Zhang
cInstitute of Cardiovascular Research, University of South China, Hunan 421001, China;
Jared Osland
aDepartment of Internal Medicine, Southern Illinois University School of Medicine, Springfield, IL, 62702;
John Gao
dDepartment of Pathology & Laboratory Medicine, Southern Illinois University School of Medicine, Springfield, IL, 62702;
Tian Zhang
eDepartment of Surgery, Southern Illinois University School of Medicine, Springfield, IL, 62702;
Stuart J. Frank
fDepartment of Medicine, University of Alabama, Birmingham, AL, 35294;
Martin A. Javors
gDepartment of Psychiatry, University of Texas Health Science Center, San Antonio, TX, 78229;
Rong Yuan
aDepartment of Internal Medicine, Southern Illinois University School of Medicine, Springfield, IL, 62702;
John J. Kopchick
hDepartment of Biomedical Sciences, Heritage College of Osteopathic Medicine, Ohio University, Athens, OH, 45701;
Liou Y. Sun
iDepartment of Biology, University of Alabama, Birmingham, AL, 35294
Jie Chen
bDepartment of Cell and Developmental Biology, University of Illinois Urbana–Champaign, Urbana, IL, 61801;
Andrzej Bartke
aDepartment of Internal Medicine, Southern Illinois University School of Medicine, Springfield, IL, 62702;
Author contributions: Y.F., L.Y.S., J.C., and A.B. designed research; Y.F., C.M.H., J.D., A.R.-O., E.A., S.M., C.Z., J.O., J.G., T.Z., and M.A.J. performed research; S.J.F., R.Y., J.J.K., and J.C. contributed new reagents/analytic tools; Y.F., C.M.H., and J.C. analyzed data; Y.F., J.D., J.C., and A.B. wrote the paper; and J.C. and A.B. provided funding.
Significance
In various animal species, including mammals, longevity can be extended by rapamycin, an inhibitor of mTOR (mechanistic target of rapamycin). mTOR acts through two complexes: mTORC1 and mTORC2. Antiaging effects of rapamycin are mediated by suppression of mTORC1, while the role of mTORC2 in aging remains to be elucidated. Here, we report that mTORC2 plays a positive role in regulating longevity via maintenance, or enhancement, of whole-body homeostasis. When mTORC2-mediated homeostasis was disrupted by rapamycin in the remarkably long-lived GHR-KO mice (in which mTORC1 signaling is low, while mTORC2 signaling is elevated), their life span was shortened. Hence, a selective approach toward mTORC1 inhibition without impairing mTORC2 is important in devising a strategy for slowing aging.
Abstract
It is well documented that inhibition of mTORC1 (defined by Raptor), a complex of mechanistic target of rapamycin (mTOR), extends life span, but less is known about the mechanisms by which mTORC2 (defined by Rictor) impacts longevity. Here, rapamycin (an inhibitor of mTOR) was used in GHR-KO (growth hormone receptor knockout) mice, which have suppressed mTORC1 and up-regulated mTORC2 signaling, to determine the effect of concurrently decreased mTORC1 and mTORC2 signaling on life span. We found that rapamycin extended life span in control normal (N) mice, whereas it had the opposite effect in GHR-KO mice. In the rapamycin-treated GHR-KO mice, mTORC2 signaling was reduced without further inhibition of mTORC1 in the liver, muscle, and s.c. fat. Glucose and lipid homeostasis were impaired, and old GHR-KO mice treated with rapamycin lost functional immune cells and had increased inflammation. In GHR-KO MEF cells, knockdown of Rictor, but not Raptor, decreased mTORC2 signaling. We conclude that drastic reduction of mTORC2 plays important roles in impaired longevity in GHR-KO mice via disruption of whole-body homeostasis.
Mechanistic target of rapamycin (mTOR) plays central roles in growth, metabolism, and aging. It acts via two distinct complexes: mTORC1 and mTORC2, defined by Raptor and Rictor, respectively (1–3). Rapamycin, an inhibitor of mTOR, inhibits mTORC1, and longer rapamycin treatment also inhibits mTORC2. Rapamycin was the first drug shown to extend longevity in a mammal (4). The effects of rapamycin on longevity were accounted for by mTORC1 inhibition, whereas information on mTORC2 is generally lacking (2). However, it seems that many of the negative adverse effects of rapamycin treatment are mediated by inhibition of mTORC2 (5). It has been recently shown that Rictor has positive effects on a variety of functions involved in whole-body homeostasis (6–10). Although at this time, the role of mTORC2 in the regulation of longevity is uncertain (11), several lines of evidence imply that mTORC2 may have opposite effects on aging compared with mTORC1. For instance, Rictor loss-of-function mutants in Caenorhabditis elegans had decreased life spans by 24–43% on a standard diet (12), muscle-specific inactivation of Rictor in mice produced glucose intolerance (13), kidney-specific knockout of Rictor reduced protection of kidneys from stress (14), and adipose-specific knockout of Rictor led to a disproportionately enlarged pancreas and hyperinsulinemia (15). Interestingly, transcriptional down-regulation of mTORC1 and transcriptional up-regulation of mTORC2 was reported to be associated with human longevity (16). It is vital to understand how mTORC2 regulates aging in a mammal. It was shown that both heterozygous deletion (Rictor+/−) and liver knockout of Rictor (l-RKO) reduced the life span of male, but not female, mice (11). However, neither protein levels of Rictor and AKT (the major downstream effector of mTORC2) nor phosphorylation of AKT, were differentially affected in male and female mice. Thus, although life span in male Rictor+/− or l-RKO mice was shortened, the connection between known mTORC2 signaling effectors and life span remains to be elucidated.
mTORC2 is a key regulator of lipid biosynthesis and AKT-mediated survival signaling (17). A recent study reported that removal of the C terminus of Avo3 (yeast homolog of Rictor) produced a rapamycin-sensitive TORC2 variant, and unlike TORC1, “rapamycin inhibition of TORC2 triggered a rapid cell-cycle arrest in G2/M” (17), indicating that rapamycin could inhibit mTORC2 if the C terminus of Rictor was manipulated by some conditions and the “rapamycin-sensitive TORC2 variant” had different downstream outputs compared with TORC1 functions. mTORC2 is regulated by growth hormone (GH)-dependent growth factors. GH is essential for growth and metabolism and is involved in the control of aging (18). It binds and signals through GH receptor (GHR). Therefore, deletion of GHR eliminates GH signaling and its biological functions. GHR-KO (GHR knockout) mice have been a valuable tool to study GH functions, including its relationship to longevity (19). GHR-KO mice are dwarf (19), extremely insulin sensitive (20), and have their life span extended up to 40% (19). Importantly, compared with their normal (N) littermates, GHR-KO and several other long-lived mice have decreased mTORC1 and increased mTORC2 signaling (21), which may play a role in their extended longevity. Therefore, we decided to examine how prolonged rapamycin treatment alters mTORC1 and mTORC2 signaling in GHR-KO mice. We were especially interested in the effects of low mTORC1 signaling in the presence of high activity of mTORC2, which could be reduced by prolonged rapamycin treatment, on the animals’ whole-body homeostasis and consequent life span. Here, we demonstrate that the benefits of low mTORC1 signaling in GHR-KO mice are diminished, which is associated with reduction of mTORC2 signaling and disruption of whole-body homeostasis.
Results
Life Span of GHR-KO Mice Was Reduced by Rapamycin Treatment.
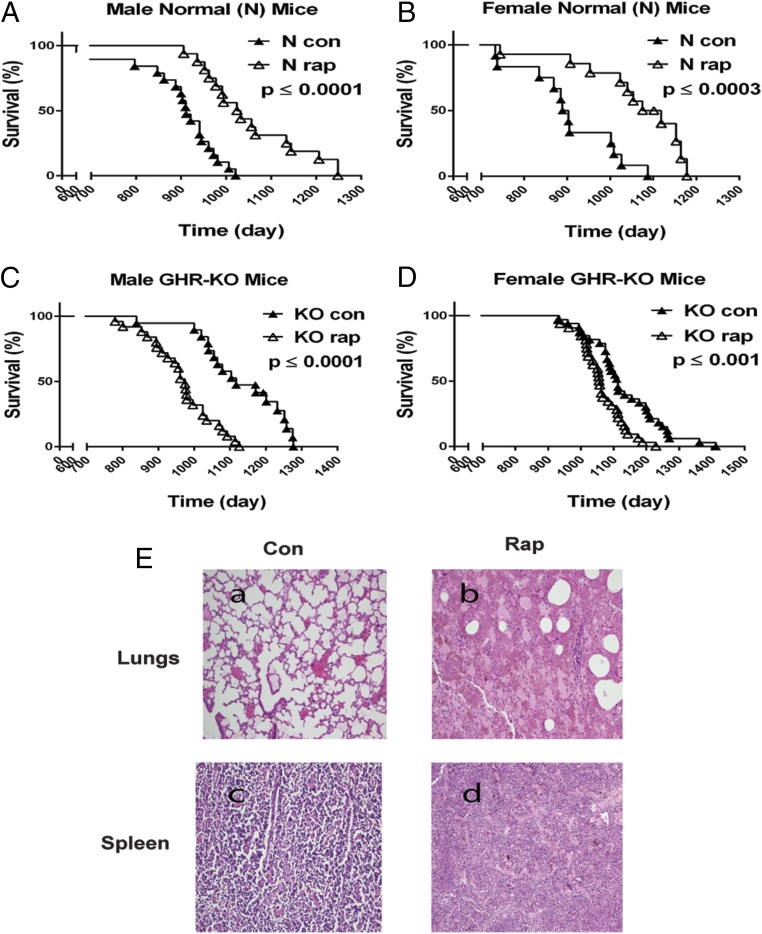
To examine the effects of prolonged rapamycin treatment on the longevity of GHR-KO mice, animals were treated with rapamycin starting at 600–700 d of age, resembling a study in which rapamycin fed late in life extended life span in genetically heterogeneous mice with intact GH signaling (4). Our longevity study was performed using the dose and injection protocol of rapamycin shown to extend the life span of C57BL/6 mice with normal GH signaling (22). As expected, prolonged rapamycin treatment extended the life span of both male and female control N mice (Fig. 1 A and B). However, the same treatment in GHR-KO mice reduced life span of both males and females (Fig. 1 C and D). Mean life span of GHR-KO for both sexes combined was shortened 9% by rapamycin treatment (P < 0.001). Rapamycin treatment had greater effects on the life span of male GHR-KO mice (12.5% shortened; P < 0.0001; Fig. 1C) compared with female GHR-KO mice (6% shortened; P < 0.001; Fig. 1D and Table 1). Maximum life span of both male and female GHR-KO mice was also decreased by rapamycin (Fig. 1 C and D and Table 1). End-of-life histology showed that GHR-KO mice with prolonged rapamycin treatment had an accumulation of pigmented alveolar macrophage-like cells in the lungs and pigmented macrophage-like cells in the spleen (Fig. 1E). Other organs examined, including the liver, heart, kidney, pancreas, and muscle, appeared normal and did not differ pathologically from GHR-KO mice treated with vehicle (Table 2).
Prolonged rapamycin treatment reduced the life span of GHR-KO mice. (A) Survival plot for male N littermates (n = 35). (B) Survival plot for female N littermates (n = 26). (C) Survival plot for male GHR-KO mice (n = 45). (D) Survival plot for female GHR-KO mice (n = 65). The mice were i.p injected with rapamycin starting at 600–700 d of age for a longevity study. P values were calculated by the log-rank test. (E) Major pathological lesions in GHR-KO mice including slides of sections (hematoxylin and eosin) from the lungs or spleen of GHR-KO mice with or without rapamycin treatment. An example of lung tissue (a) or spleen (c) from GHR-KO mice treated with vehicle. An example of accumulation of pigmented macrophage-like cells in the lungs (b) or spleen (d) from the mice treated with rapamycin (n = 3–5 for control, n = 3–5 for rapamycin). P values were calculated between rapamycin-treated and vehicle-treated groups.
Table 1.
Analysis of life span for the rapamycin-treated GHR-KO mice
| Sex and Treatment | n | Life span, d | Median | Survival beyond 3 y, % |
| Male | ||||
| Rapamycin | 20 | 975 ± 96**** | 488 | 0 |
| Control (vehicle) | 25 | 1,117 ± 85 | 558 | 38 |
| Female | ||||
| Rapamycin | 32 | 1,056 ± 53*** | 528 | 30 |
| Control (vehicle) | 33 | 1,113 ± 90 | 556 | 57 |
| Combined | ||||
| Rapamycin | 52 | 1,024 ± 77**** | 512 | 20 |
| Control (vehicle) | 58 | 1,113 ± 81 | 556 | 55 |
Data represented as mean ± SEM. P values were calculated by a log-rank test followed by a Tukey post hoc analysis between rapamycin treated and control groups.
*P ≤ 0.05.
**P ≤ 0.01.
***P ≤ 0.001.
****P ≤ 0.0001.
Table 2.
End-of-life histology study
| Organ | Control (vehicle) | Rapamycin |
| Heart | Normal | Normal |
| Lung | Mild pigmented macrophage-like cells | Numerous nonpigmented and pigmented macrophage-like cells; some of lung had hyphae |
| Liver | Minimal steatosis with minimal hemosiderin deposition in Kupffer cells | Minimal steatosis with minimal hemosiderin deposition in Kupffer cells |
| Kidney | Normal | Normal |
| Spleen | Mild hemosiderin deposition | Moderate hemosiderin deposition |
| Pancreas | Normal | Normal |
| Muscle | Normal | Normal |
Observations were based on three to five mice for each group.
Prolonged Rapamycin Treatment in GHR-KO Mice Did Not Change mTORC1 but Decreased mTORC2 Signaling in Multiple Tissues.
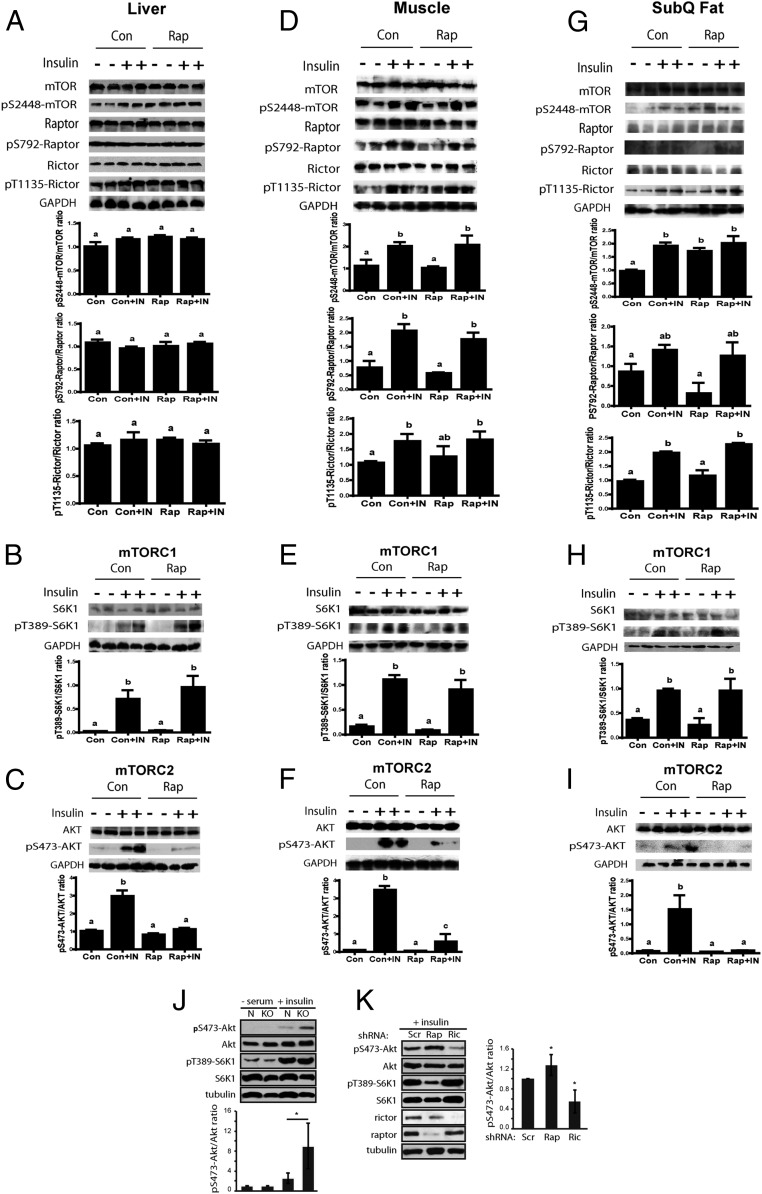
The life span of GHR-KO mice was reduced by prolonged rapamycin treatment. To better understand this unexpected finding, we evaluated how prolonged rapamycin treatment affected mTOR signaling in these animals. Young adult GHR-KO mice have been reported to have suppressed mTORC1 and elevated mTORC2 signaling in several tissues, including the liver (21). Therefore, mTOR signaling was examined in GHR-KO mice treated with rapamycin starting at 3 mo of age for 20 wk. Western blot analyses of liver samples showed that the levels of pS2448-mTOR, pS792-Raptor, and pT389-S6K1 (an mTORC1 substrate) were not significantly different between GHR-KO mice treated with rapamycin or vehicle, indicating that mTORC1 signaling was unaltered in the liver of these mice (Fig. 2 A and B). The levels of pT1135-Rictor were similar to the controls treated with vehicle (Fig. 2A), but phosphorylation of pS473-AKT (an mTORC2 substrate) decreased significantly after 20 wk of rapamycin treatment (Fig. 2C). These results were opposite to the hepatic response of the N littermates to identical treatment (23). Unexpectedly, the data showed that rapamycin did not significantly inhibit mTORC1 signaling in the liver of GHR-KO mice in the present study. In addition, the Western blot analyses of muscle and s.c. adipose tissue were performed to examine mTOR signaling in these two tissues. The data from these two tissues consistently demonstrated that mTORC1 signaling was not drastically altered by prolonged (20 wk) rapamycin treatment in GHR-KO mice (Fig. 2 D, E, G, and H), whereas mTORC2 signaling was significantly reduced (Fig. 2 F and I). Thus, in the absence of GH inputs in GHR-KO mice, prolonged rapamycin treatment reduced the signaling of mTORC2 without significantly affecting mTORC1.
Analyses of mTOR signaling in the liver, muscle, and s.c. adipose tissue of GHR-KO mice with rapamycin treatment for 20 wk, and MEF cells from GHR-KO or N mice with shRNA Knockdown of Raptor or Rictor. (A–C) Hepatic mTOR signaling. (D–F) mTOR signaling in the muscle. (G–I) mTOR signaling in s.c. adipose tissue. (J) Insulin-stimulated mTOR signaling in MEF cells. (K) mTOR signaling in KO MEF cells with shRNA knockdown of Raptor or Rictor. All bar graphs represent the mean ± SEM of three experiments, from a total of six to seven mice. P values were calculated between rapamycin-treated and vehicle-treated groups. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
To directly assess the roles of mTORC2 versus mTORC1 in the observed signaling alterations in vivo, we took advantage of MEF cells derived from the mice in our study. GHR-KO MEF cells displayed increased pS473-AKT in response to insulin stimulation (Fig. 2J), which was impaired by the knockdown of Rictor (Fig. 2K), thus confirming a role of mTORC2, consistent with the implications of our observations in whole animal. Knockdown of Raptor in KO MEF cells reduced mTORC1 signaling while increasing mTORC2 signaling (Fig. 2K), most likely through a well-established feedback mechanism (3). Together, our in vivo and in vitro results showed that long-lived GHR-KO mice have increased mTORC2 signaling that is inhibited by prolonged rapamycin treatment.
Insulin Sensitivity Was Impaired in Rapamycin-Treated GHR-KO Mice.
Prolonged rapamycin treatment decreased life span of GHR-KO mice and reduced signaling of mTORC2 downstream targets. These targets are vital regulators of glucose and lipid metabolism (1) and play a role in the defense against bacterial pathogens, bacterial clearance, inflammation, and wound healing (24), which are all important to maintain whole-body homeostasis. Thus, we wanted to examine whole-body homeostasis reflected by body composition and glucose, lipid, and energy metabolism, as well as the immune system.
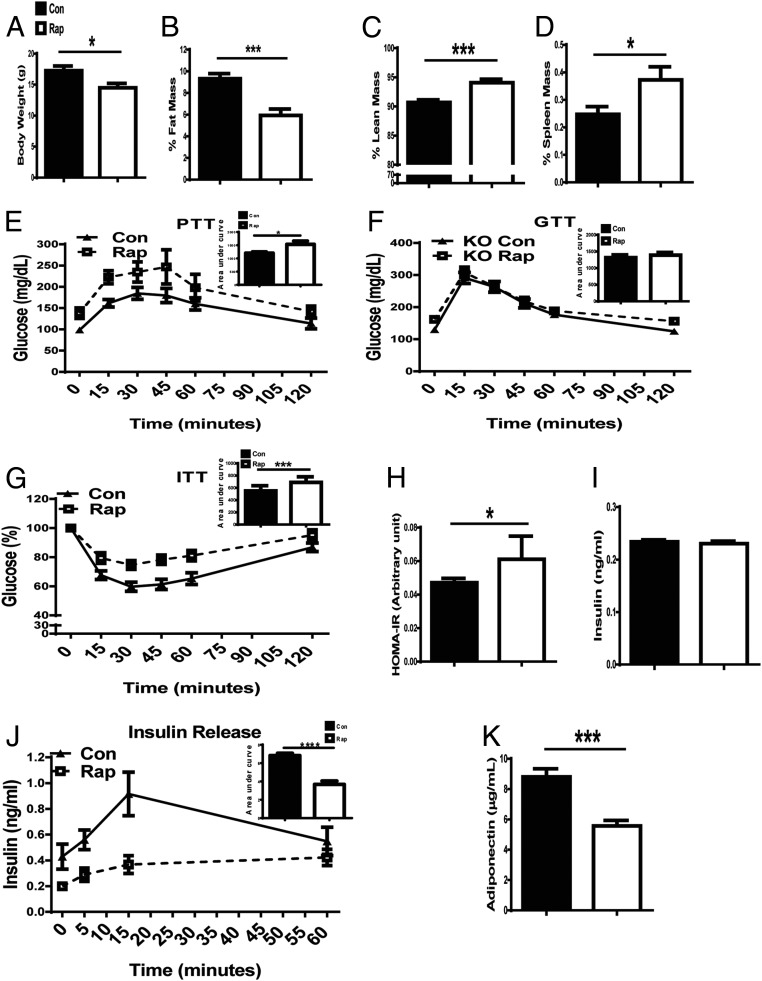
In GHR-KO mice injected with rapamycin for 20 wk, body weight (BW; Fig. 3A) and percentage of fat mass (Fig. 3B) were lower than in vehicle-injected GHR-KO controls, whereas percentage of lean mass (Fig. 3C) and relative spleen weight (Fig. 3D) were higher. Glucose homeostasis is critical to longevity and has been shown to be affected by rapamycin treatment in various mouse models with normal GH action (23, 25). GHR-KO mice are hypoinsulinemic and extremely insulin sensitive (20). Hence, it was important to examine how rapamycin affected glucose homeostasis in GHR-KO mice. Prolonged rapamycin treatment elevated fasting glucose levels, possibly as a result of increased gluconeogenesis [assessed by a pyruvate tolerance test (PTT)] (Fig. 3E). Glucose tolerance did not differ from that measured in GHR-KO mice treated with vehicle (Fig. 3F), but the mice treated with rapamycin became insulin resistant (Fig. 3 G and H). Insulin sensitivity is affected by the circulating levels of insulin (basal and stimulated). In the current study, basal circulating insulin levels were not altered by 20 wk of rapamycin treatment (Fig. 3I), and yet after 20 wk of rapamycin treatment, the mice became insulin resistant, which may be related to reduced levels of adiponectin (an insulin sensitizer; Fig. 3K) and a slower glucose-stimulated insulin release at already-low levels of insulin in GHR-KO mice (Fig. 3J). These data show that after 20 wk of rapamycin treatment, insulin sensitivity in GHR-KO mice was decreased, and the animals became insulin resistant, which could relate to reduced adiponectin levels. Thus, in the absence of GH action, prolonged rapamycin treatment induced detrimental alterations in glucose homeostasis, which were different from the observation in N littermates given an identical treatment (23).
Prolonged rapamycin treatment impaired glucose homeostasis in GHR-KO mice. (A) BW. (B) Relative fat mass [absolute value (g)/BW (g)] × 100; total fat includes s.c. fat from the thighs, a pair of epididymal depots, a pair of perirenal depots, and interscapular brown fat. (C) Relative lean mass. (D) Relative mass of the spleen. (E) PTT: 16-h fasted mice underwent PTT by i.p. injection with 1.5 g sodium pyruvate per kilogram BW. (F) GTT: 16-h fasted mice underwent GTT by i.p. injection with 1 g glucose per kilogram BW. (G) ITT: mice were injected i.p. with 1 IU/kg BW porcine insulin. (H) HOMA-IR (homeostatic model for assessment of insulin resistance): HOMA-IR = (FPI × FPG)/22.5, where FPI is fasting plasma insulin and FPG is fasting plasma glucose. (I) Plasma insulin levels. (J) Insulin releasing: 16-h fasted mice were injected i.p. with 1 g glucose per kilogram BW, and then blood was taken from the tail vein and plasma insulin was measured by ELISA. (K) Plasma adiponectin levels: the blood was taken at killing from mice fasted for 16 h and measured by ELISA. Data were represented as the mean ± SEM (n = 8–16 for control, n = 9–16 for rapamycin). P values were calculated between rapamycin-treated and vehicle-treated groups. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
Circulating Levels of Cholesterol, Triglycerides, and Free Fatty Acids Were Elevated in Rapamycin-Treated GHR-KO Mice.
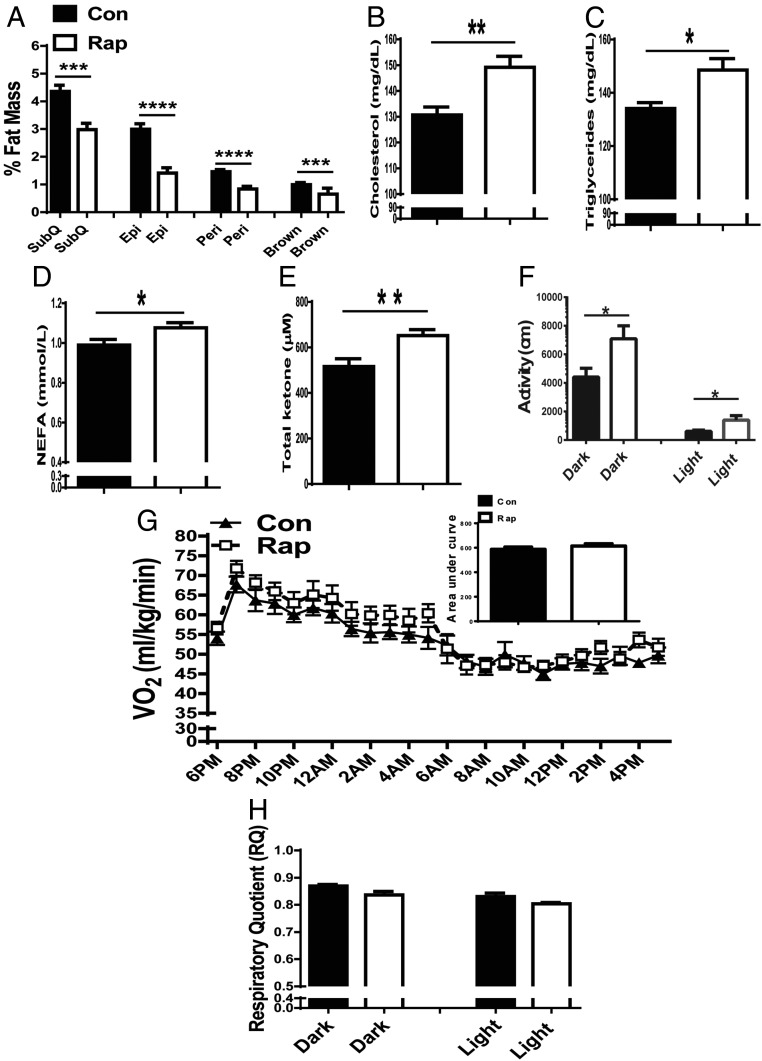
GHR-KO mice have enlarged, but functionally unique and “healthy,” intra-abdominal adipose tissue that has down-regulated IL-6 and up-regulated insulin receptor and adiponectin compared with their N littermates. Removal of abdominal fat led to detrimental effects in GHR-KO mice (26). Twenty weeks of rapamycin treatment reduced adiposity in GHR-KO mice in the current study. Thus, it was of interest to know how lipid homeostasis and the related energy metabolism in GHR-KO mice was affected. After 20 wk of rapamycin treatment, the main white fat depots including s.c., epididymal and perirenal, as well as interscapular brown fat, in GHR-KO mice were significantly decreased (Fig. 4A), likely accounting for the increase in circulating levels of cholesterol (Fig. 4B), triglycerides (Fig. 4C), and free fatty acids (Fig. 4D). Total circulating ketone bodies were also elevated (Fig. 4E). Unlike the alterations in adipose tissue, the increase in relative lean mass (Fig. 3C) by prolonged rapamycin treatment may promote an increase in glucose usage, and possibly lead to increased spontaneous locomotor activity. Indeed, spontaneous locomotor activity was increased in GHR-KO mice treated with rapamycin (Fig. 4F). Despite the altered lipid profile, GHR-KO mice treated with rapamycin maintained their normal metabolic rate, reflected by similar levels of oxygen consumption (Fig. 4G) and respiratory quotient (Fig. 4H). Taken together, rapamycin-treated GHR-KO mice lost much of their functionally unique and “healthy” fat depots, which was accompanied by detrimental alterations in their circulating lipid profile. However, energy metabolism parameters remained stable, and some measurably beneficial responses occurred, such as increased spontaneous locomotor activity and increased circulating levels of total ketone bodies.
Prolonged rapamycin treatment resulted in detrimental lipid profiles in GHR-KO mice. (A) Relative mass of individual fat depots: s.c. (SubQ) fat from the thighs, a pair of epididymal depots (Epi) fat, a pair of perirenal depots (Peri) fat, and interscapular brown (Brown) fat. (B) Plasma cholesterol levels. (C) Plasma triglyceride levels. (D) Plasma nonesterified free fatty acid levels. (E) Plasma total ketone bodies. (F) Spontaneous locomotor activity. (G) Oxygen consumption VO2 (mL/kg/min). (H) Respiration quotient (RQ). (F–H) measured using indirect calorimetry (AccuScan Instruments). Data represented as the mean ± SEM (n = 8–16 for control, n = 9–16 for rapamycin). P values were calculated between rapamycin-treated and vehicle-treated groups. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001.
Effects of Rapamycin on Some Aspects of Immune System and Inflammation Status in GHR-KO Mice.
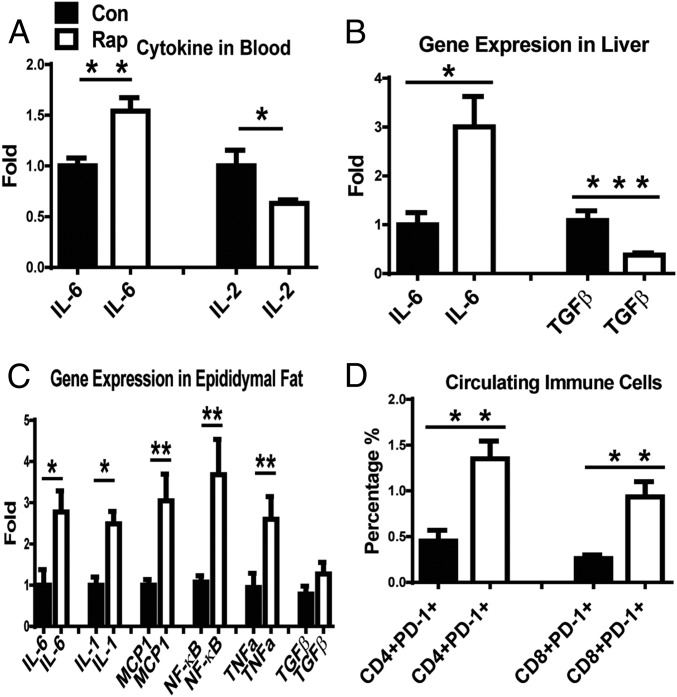
Long-lived GHR-KO mice with decreased mTORC1 and increased mTORC2 signaling have significantly lower disease burden and a slower age-related accumulation of various pathological changes (27). They also have lower expression of inflammatory genes in adipose tissue than their N littermates (26). The current study showed that mTORC2 signaling was drastically reduced by prolonged rapamycin treatment. Therefore, it is possible that mTORC2-mediated immune function/status could have been altered by this treatment. Indeed, GHR-KO mice with prolonged rapamycin treatment had higher circulating levels of IL-6 and lower levels of IL-2 compared with GHR-KO mice treated with vehicle (Fig. 5A). The expression of pro- and antiinflammatory genes was altered in the liver, particularly in epididymal fat (Fig. 5 B and C). In rapamycin-treated GHR-KO mice, the hepatic expression of TGFβ (an antiinflammatory gene) was reduced (Fig. 5B), and the expression of some proinflammatory genes was elevated in epididymal fat (Fig. 5C).
Effects of rapamycin on the immune system and inflammation status in GHR-KO mice. (A) Plasma levels of IL-6 and IL-2. (B and C) Altered gene expression of pro- and antiinflammation in the liver (B) or in epididymal fat depot (C). (D) Increase in percentage of PD-1–positive CD4 and CD8 cells in the blood from old GHR-KO mice treated with rapamycin for 10 mo. Data represented as the mean ± SEM (n = 5–9 for control, n = 5–12 for rapamycin). P values were calculated between rapamycin-treated and vehicle-treated groups. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001 for all panels.
It was reported that in older adult humans, 6 wk of treatment with a rapamycin analog enhanced immune function by reducing circulating CD4 and CD8 cells positive for PD-1 (programmed death-1, which negatively regulates immune function) (28). To examine the impact of prolonged rapamycin treatment on CD4- and CD8-mediated immune function in old GHR-KO mice, a similar study was performed by injecting GHR-KO mice with rapamycin starting at 600 d of age for 10 mo. The blood was used for immune staining for PD-1–positive CD4 and CD8 cells. In contrast to the results reported in older adult humans (28), prolonged rapamycin treatment significantly increased the percentage of circulating PD-1–positive CD4 and CD8 T cells in old rapamycin-treated GHR-KO mice (Fig. 5D), indicating that the immune functions mediated by CD8 and CD4 cells may have been reduced in these animals. The present data on inflammation-related gene expression and certain subsets of CD4 and CD8 T cells represent preliminary assessment of the immune function/condition. Systematic detailed studies of the immune functions in GHR-KO mice with prolonged rapamycin treatment are of interest for future work. However, present findings suggest that prolonged rapamycin treatment led to some groups of circulating immune cells (CD4- and CD8-positive cells) in GHR-KO mice losing their immune function by entering PD-1–positive status. Also, some proinflammatory genes were up-regulated, and some antiinflammatory genes were down-regulated in these mice.
Discussion
In long-lived GHR-KO mice, prolonged rapamycin treatment did not further extend, but unexpectedly shortened, life span. One possible reason could be that prolonged rapamycin treatment further decreases the already low levels of mTORC1 signaling in these animals, which could adversely affect the benefits of mTORC1 inhibition. However, mTORC1 signaling was not further reduced in three key metabolic tissues of GHR-KO mice with prolonged rapamycin treatment compared with GHR-KO mice treated with vehicle (Fig. 2). We cannot explain why prolonged rapamycin treatment did not further decrease mTORC1 signaling in these animals, and also cannot rule out the possibility that mTORC1 signaling may have been further reduced in other tissues. In contrast, a significant reduction of mTORC2 signaling was evident in each of the examined tissues of GHR-KO mice treated with rapamycin (Fig. 2). Decreased mTORC2 signaling and impaired whole-body homeostasis (which could result from reduced mTORC2 signaling) in rapamycin-treated GHR-KO mice might have contributed to the effect of prolonged rapamycin treatment on the life span of GHR-KO mice. Thus, our data indicated that mTORC2 may play a beneficial role in longevity via improving or maintaining whole-body homeostasis. Based on our data and data from previous studies (6–11), we propose the following concept: if whole-body homeostasis is impaired (which was associated with the significant reduction of mTORC2 in our study), life span could be shortened, and if mTORC2 signaling is unaltered or enhanced, inhibition of mTORC1 will lead to extension of life span. The effects of altered mTOR signaling on longevity would thus reflect a balance between inhibition of mTORC1 and enhancement, or maintenance, of mTORC2.
Because of the lack of a specific mTORC2 inhibitor, rapamycin was used as a tool in this special mouse model (GHR-KO mice that have extended life span with suppressed mTORC1 and elevated mTORC2 signaling) to determine whether reducing mTORC2 signaling in the animals, in which the activity of mTORC1 is already low, can affect the life span. The results revealed that rapamycin extended the life span of control N mice, as expected from previous studies (4, 22), including results of deleting the mTORC1 target, S6K1 (29), whereas the same treatment in GHR-KO mice had the opposite effect. We suspect that reduced longevity of GHR-KO mice by prolonged rapamycin treatment may have been a result of disruption of whole-body homeostasis. This possibility was addressed from several angles. First, several indications of whole-body homeostasis, such as glucose and lipid metabolism, and some aspects of the immune system were impaired in these unique mice after treatment with rapamycin. Second, mTORC2 signaling was drastically decreased in tissues from GHR-KO mice after rapamycin treatment, and higher mTORC2 signaling in KO MEF cells was reduced by the knockdown of Rictor, but not by the knockdown of Raptor. Third, after rapamycin treatment, old GHR-KO mice lost more functional immune cells and had elevated expression of some proinflammatory genes. Finally, the life span of GHR-KO mice was decreased after rapamycin treatment.
The role of mTORC1 signaling in longevity has been the focus of previous studies; information on the possible role of mTORC2, and the balance between mTORC1 and mTORC2 in the control of longevity, is very limited. Although rapamycin is not the ideal pharmacological drug to distinguish the function of mTORC1 and mTORC2, we decided to use it because of the lack of a specific mTORC2 inhibitor and limited information from the mice with global genetic reduction of Rictor (Rictor+/−) or genetic knockout of Rictor in the liver (l-RKO mice) (11). Our study sheds light on a particular aspect of the role of mTOR signaling in longevity; namely, the balance between inhibition of mTORC1 and enhancement, or maintenance, of mTORC2. In GHR-KO mice, prolonged rapamycin treatment decreased mTORC2 signaling, and mTORC1 signaling was not profoundly changed, thus leading to the altered balance between these two complexes and associating with disrupted whole-body homeostasis and reduced life span. This change is different from what was observed in N littermates of GHR-KO mice (23). Interestingly, similar alterations were reported in long-lived mtor+/− mlst8+/− female mice, in which the Raptor/mTOR ratio was decreased but Rictor/mTOR ratio was similar to those measured in wild-type mice (30). Taken together, our findings support the concept that mTORC1 and mTORC2 signaling have distinctive functions in the control of longevity and that mTORC2 plays a positive role in longevity via maintenance of whole-body homeostasis. Hence, a selective approach toward mTORC1 inhibition (without impairing mTORC2) may be important in devising a strategy for slowing aging.
Materials and Methods
Mice Maintenance and Rapamycin Injection.
All animal procedures were approved by the Laboratory Animal Care and Use Committee of Southern Illinois University School of Medicine. Mice were housed under temperature- and light-controlled conditions (23 °C ± 1 °C, 12 h light/12 h dark cycle) with ad libitum access to food (Chow 5001 with 23.4% protein, 5% fat, and 5.8% crude fiber; LabDiet PMI Feeds) and water. Our breeding colony was developed by crossing GHR-KO males with 129 Ola/BALB/c background generated in the J.J.K. laboratory (19) with N females derived from crossing of C57BL/6 and C3H strains, and thereafter breeding of the resulting animals in a closed colony without brother × sister mating. Thus, the animals have a heterogeneous genetic background. Starting at 3 mo of age, male GHR-KO mice were injected intraperitoneally (i.p.) with 4 mg/kg BW rapamycin (LC Laboratories) or vehicle every other day, according to a previously established protocol (22), and were killed after 20 wk of rapamycin treatment. For the longevity study, both male and female GHR-KO or N mice starting at 600–700 d of age were injected with rapamycin or vehicle, using the same protocol as stated earlier (22).
PTT, Glucose Tolerance Test, Insulin Tolerance Test, and Insulin Releasing Experiments.
Sixteen-hour-fasted mice underwent PTT by i.p. injection with 1.5 g sodium pyruvate per kilogram of BW, or underwent glucose tolerance test (GTT) by i.p. injection with 1 g glucose per kilogram of BW. Blood glucose levels were measured at 0, 15, 30, 45, 60, and 120 min with a PRESTO glucometer (AgaMatrix). Nonfasted mice were injected i.p. with 1 IU porcine insulin (Sigma) per kilogram of BW. Blood glucose levels were measured at 0, 15, 30, 45, 60, and 120 min for insulin tolerance test (ITT). The data for GTT and PTT are presented as absolute value, and for ITT are presented as a percentage of baseline glucose. Sixteen-hour fasted mice underwent insulin-releasing experiments by injecting 1 g glucose i.p. per kilogram of BW, and blood was taken from the tail vein at 0, 5, 15, and 60 min for insulin measurement.
Indirect Calorimetry.
Mice injected with rapamycin for 20 wk were subjected to indirect calorimetry (AccuScan Instruments). The system uses zirconia, infrared sensors, and light beam arrays to monitor oxygen consumption (VO2), carbon dioxide output (VCO2), and spontaneous locomotor activity inside respiratory chambers in which individual mice were tested. After 24 h of acclimation, mice were monitored in the chambers for 24 h with ad libitum access to food (Chow 5001; LabDiet PMI Feeds) and water. All comparisons are based on animals studied simultaneously in eight different chambers connected to the same O2, CO2, and light beam sensors. Gas samples were collected and analyzed every 10 min per animal, and the data were averaged for each hour.
Assessment of Blood Chemistry.
After 20 wk of rapamycin treatment, mice were fasted for 16 h (overnight) and killed the next morning. At the time they were killed, plasma was collected at fasting condition by cardiac puncture. The blood was mixed with EDTA, followed by centrifugation at 10,000 × g for 15 min at 4 °C for plasma collection. Per the manufacturer’s protocol, insulin was measured with a Mouse Insulin ELISA Kit (Crystal Chem), adiponectin with a Mouse Adiponectin ELISA Kit (Linco Research), IL-6 with a Mouse Interleukin‐6 ELISA MAX Deluxe Kit (BioLegend, Inc), and IL-2 with a Mouse Cytokine Chemi Demo Kit (Quansys Biosciences), respectively. Total ketone bodies and nonesterified free fatty acids were measured via colorimetric assays from Wako Chemicals, and cholesterol and triglycerides were measured with kits from Pointe Scientific.
Mouse Embryonic Fibroblast Isolation and Culture.
MEF cells were isolated from GHR-KO and N mice. In brief, E10-10.5 embryos were collected and triturated in DMEM, and then plated in DMEM with 10% FBS in 5% CO2. Cells were passed and replated at constant density (1.5 × 104/cm2) every 3–4 d. Spontaneously immortalized MEF cell lines were generated from the cultures. MEF cells, derived from N or GHR-KO mice, were maintained in DMEM (4.5 g/L glucose) supplemented with 10% FBS and 1% penicillin-streptomycin at 37 °C with 5% CO2. shRNAs for Raptor, Rictor, and a negative control (scrambled hairpin sequence) in the pLKO.1-puro vector were purchased from Sigma-Aldrich (MISSION TRC), as previously characterized (31). The clone IDs used were shRaptor, {"type":"entrez-nucleotide","attrs":{"text":"NM_028898.1","term_id":"30061324","term_text":"NM_028898.1"}}NM_028898.1–3729s1c1; shRictor, {"type":"entrez-nucleotide","attrs":{"text":"NM_030168.2","term_id":"46048255","term_text":"NM_030168.2"}}NM_030168.2–6240s1c1. Lentivirus packaging and viral transduction of cells were performed as previously described (32).
Western Blot Analysis.
Mice were fasted for 16 h. Half of the animals from each experimental group at fasting condition were used to collect plasma. Another half of the animals from each experimental group were used to collect the tissues by injecting the mice with insulin or saline through the liver portal vein to stimulate the insulin signaling pathway, according to a previously described protocol (20). Two minutes after insulin injection, mice were killed and the tissues were collected. Approximately 500-mg tissue samples were homogenized in 0.5 mL ice-cold T-PER tissue protein extraction buffer (Thermo Scientific) with protease and phosphatase inhibitors (Sigma). Total protein (40 mg) was separated by SDS/PAGE with Criterion XT Precast Gel (Bio-Rad) and blotted with antibodies. Antibodies, including mTOR, phospho-mTOR (Ser2448), Raptor, phospho-Raptor (Ser792), Rictor, phospho-Rictor (Thr1135), S6K1, phospho-S6K1 (Thr389), AKT, phospho-AKT (Ser473), and GAPDH, were obtained from Cell Signaling Technology. Western blots were quantified with Multi Gauge V3.0 software (Fujifilm North America).
Immunostaining and Flow Cytometry.
Peripheral blood mononuclear cells were isolated from the whole blood by density gradient centrifugation, according to a previously established protocol (33). Whole blood was collected from nonfasting male GHR-KO mice treated with rapamycin, as described earlier, for 10 mo, starting at around 600 d of age. Peripheral blood mononuclear cells were harvested by centrifuging at 300 × g at room temperature for 15 min, and washed twice with ice-cold PBS with 2% FBS. The cells were resuspended in PBS with 2% BSA with ≥90% viability. The cell suspension was aliquoted in a tube with 4 × 105 cells for immune staining with anti-CD4 FITC, or anti-CD8 FITC, or combined with anti–PD-1 PE (BD Biosciences) for 45 min on ice. After one wash with PBS with 2% BSA, the cells with 500 µL PBS were used for flow cytometry. One hundred thousand events were collected on Accuri C6 (BD Biosciences).
RT-PCR.
mRNA expression was analyzed by quantitative RT-PCR, using the StepOne System and SYBR Green MasterMix (Applied Biosystems). RNA was extracted using an RNeasy mini kit or RNeasy Lipid Tissue Mini Kit (Qiagen), following the manufacturer’s instructions. Relative expression was calculated as previously described (26).
End-of-Life Pathological Analysis.
A detailed pathological evaluation was conducted on the mice used in the life span study when they died. Organs and tissues were preserved in a neutral 10% buffered formalin, and the fixed tissues were processed conventionally, embedded in paraffin, sectioned at 3 µm, and stained with hematoxylin and eosin. Diagnosis of each histopathological change and severity of major lesions were determined with histological classifications in aging mice, as previously described (27).
Statistical Analysis.
Data are presented as means ± SEM. Differences between two groups were assessed with unpaired two-tailed Student’s t tests. A log-rank test followed by a Tukey post hoc analysis between rapamycin-treated and control groups was used for a longevity study. All statistical analyses and graphs were performed using Prism 6 (GraphPad Inc.).
Acknowledgments
We thank Dr. Richard A. Miller and Dr. Z. David Sharp for their discussion. We would also like to thank Alexander Wang and Kelly Zhang for experimental assistance. This study was supported by grants from the National Institutes of Health [AG019899, AG038850, AG031736, and AG051869 (to A.B.), and AR048914 and GM089771 (to J.C.)].
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1717065115/-/DCSupplemental.