- Journal List
- J Gerontol A Biol Sci Med Sci
- PMC6001896

Deletion of Nrip1 Extends Female Mice Longevity, Increases Autophagy, and Delays Cell Senescence
Jinyu Wang
1Department of Physiology and Pathophysiology, Peking University Health Science Center, Beijing, P. R. China
2Department of Internal Medicine, Division of Geriatrics Research, Southern Illinois University School of Medicine, Springfield
Xundi Chen
2Department of Internal Medicine, Division of Geriatrics Research, Southern Illinois University School of Medicine, Springfield
3Department of Molecular Biology, Microbiology and Biochemistry, Southern Illinois University School of Medicine, Springfield
Jared Osland
2Department of Internal Medicine, Division of Geriatrics Research, Southern Illinois University School of Medicine, Springfield
Skyler J Gerber
2Department of Internal Medicine, Division of Geriatrics Research, Southern Illinois University School of Medicine, Springfield
3Department of Molecular Biology, Microbiology and Biochemistry, Southern Illinois University School of Medicine, Springfield
Chao Luan
2Department of Internal Medicine, Division of Geriatrics Research, Southern Illinois University School of Medicine, Springfield
4Chinese Academy of Medical Sciences and Peking Union Medical College, Jiangsu Key Laboratory of Molecular Biology for Skin Diseases and STIs, Institute of Dermatology Nanjing, P. R. China
Kristin Delfino
5Department of Surgery, Center for Clinical Research, Southern Illinois University School of Medicine, Springfield
Rong Yuan
2Department of Internal Medicine, Division of Geriatrics Research, Southern Illinois University School of Medicine, Springfield
Associated Data
- Supplementary Materials
- Supplement Tables.GUID: AC88E4FC-47EC-4C22-89C3-F0587374E510Legends of Supplement Figures.GUID: 68AA515D-33A5-4308-B4E8-3E3A9B88C433
Abstract
Using age of female sexual maturation as a biomarker, we previously identified nuclear receptor interacting protein 1 (Nrip1) as a candidate gene that may regulate aging and longevity. In the current report, we found that the deletion of Nrip1 can significantly extend longevity of female mice (log-rank test, p = .0004). We also found that Nrip1 expression is altered differently in various tissues during aging and under diet restriction. Remarkably, Nrip1 expression is elevated with aging in visceral white adipose tissue (WAT), but significantly reduced after 4 months of diet restriction. However, in gastrocnemius muscle, Nrip1 expression is significantly upregulated after the diet restriction. In mouse embryonic fibroblasts, we found that the deletion of Nrip1 can suppress fibroblast proliferation, enhance autophagy under normal culture or amino acid starvation conditions, as well as delay oxidative and replicative senescence. Importantly, in WAT of old animals, the deletion of the Nrip could significantly upregulate autophagy and reduce the number of senescent cells. These results suggest that deleting Nrip1 can extend female longevity, but tissue-specific deletion may have varying effects on health span. The deletion of Nrip1 in WAT may delay senescence in WAT and extend health span.
It is well documented that there is a trade-off relationship between female sexual maturation (FSM) and longevity in wild animals across species and in laboratory animals. In our previous studies, we found and verified that the mouse inbred strains with delayed FSM and lower circulating insulin growth factor 1 (IGF1) at a young age have extended longevity (1,2). A genetic hypothesis of this trade-off suggests that, given adequate nutrition, mechanisms accelerating FSM would be favored in spite of reduced longevity and increased risks for metabolic disorders, including diabetes and obesity. This hypothesis is supported by studies of animals under calorie restriction or insulin/IGF signaling suppression, both of which have been found to delay FSM, improve metabolism, and extend longevity in a variety of species (3–5). In humans, age at menarche has been found to be significantly associated with health conditions later in life, including reduced glucose tolerance and insulin sensitivity (6). These studies suggested that using FSM as a biomarker, it is possible to identify aging-related genes. Using genetic and bioinformatics methods in studies employing mice, we identified nuclear receptor interacting protein 1 (NRIP1, also named RIP140) as a potential mediator for this trade-off relationship. Previous studies reported that Nrip1-deficient female mice have significantly delayed maturation (2,7) and are infertile (8). Therefore, it is firmly established that suppressing NRIP1 reduces female fecundity. Across mouse inbred strains, haplotype analysis found that the haplotype of Nrip1, which is associated with delayed FSM, is also associated with extended longevity (2). These lines of evidence support the hypothesis that manipulating Nrip1 may alter the longevity of mice.
NRIP1 has broad interactions with nuclear receptors, such as estrogen receptor, androgen receptor, retinoid X receptor, and liver X receptor. Its molecular effects influence regulation of the expression of clock genes (9), cytokine secretion, cell proliferation, and apoptosis (reviewed in ref. (10)). Nrip1-deficient mice are lean and have improved glucose tolerance and insulin sensitivity (10,11). These changes are remarkably similar to those found in animals that are under calorie restriction or have suppressed insulin/IGF signaling (3–5). In the current study, we compared the longevity of Nrip1 homozygous (Nrip1−/−, Hom) and heterozygous (Nrip1−/+, Het) knockout (KO) mice to wild-type (Wild) mice. To further investigate its role in the regulation of aging, we measured Nrip1 expression in different tissues at young (6 months), middle (12 months), and old (18 months) ages for both sexes. We also observed the alterations of Nrip1 expression in different tissues caused by diet restriction in female mice. We established fibroblast lines from the Hom and Het Nrip1 KO, as well as Wild embryos, to investigate the potential role of Nrip1 in autophagy and cellular senescence, which are two important mechanisms in regulating aging.
Materials and Methods
Mice
Three mouse strains were used in this project, C57BL/6J (B6) purchased from The Jackson Laboratory (TJL), global knockout Nrip1 provided by Dr. M. Parker (MP-Nrip1 KO) (2), as well as our newly developed Nrip1 global knockout strain (RY-Nrip1 KO) created by using the Cre/LoxP strategy. MP-Nrip1 KO mice used in the life-span study were generated and maintained in TJL. Mice used for other studies in this report were housed at Southern Illinois University School of Medicine (SIUSOM). The housing conditions of the two institutions have been fully described previously (1,12). Animal care and handling were conducted in accordance with NIH guidelines and the policies of TJL and SIUSOM Animal Care and Use Committee.
MP-Nrip1 mice for lifes-pan study were generated by eight breeding pairs of heterozygous females and males (MP-Nrip1−/+ × MP-Nrip1−/+). After being weaned, 144 mice that were produced between September 19, 2011 and March 11, 2012 were entered in the life-span study. The numbers of different genotypes are listed in Table 1. Genotypes of the mice were determined as we described previously (2).
Table 1.
Longevity Parameters of MP-Nrip1 Mice
| Sex | Genotype | No. | Mean (days) | SE | Median (days, 95% CI) | Age (days) | Longest 20% (days) | ||
|---|---|---|---|---|---|---|---|---|---|
| 25% Death | 75% Death | Mean | SE | ||||||
| Female | Wild | 24 | 767.0 | 34.6 | 776 (680–901) | 677 | 922 | 954.5 | 15.4 |
| Het | 37 | 898.5 | 25.3 | 958 (818–983) | 779 | 1,017 | 1,083a | 16.1 | |
| Hom | 11 | 853.4 | 56.3 | 920 (563–972) | 810 | 972 | NA | NA | |
| Male | Wild | 29 | 879.8 | 22.9 | 931 (781–978) | 773 | 981 | 1,017.8 | 9.7 |
| Het | 32 | 881.6 | 24.3 | 899 (852–945) | 824 | 962 | 1,026.2 | 16.2 | |
| Hom | 11 | 799.2 | 69.5 | 813 (749–1,000) | 761 | 1,000 | NA | NA | |
Note: CI = confidence interval; NA = not applicable.
RY-Nrip1 mice were generated using the Cre/LoxP strategy. The details for generating mice that carry Nrip1 null allele are described in Supplementary Material. Briefly, two 500-bp PCR products of Nrip1 were subcloned into a retrieval plasmid. This plasmid was then co-electroporated into recombination competent Escherichia coli with a BAC RP23 clone carrying a 204-kb insert containing the entire Nrip1 gene locus. A 15-kb genomic DNA fragment was retrieved from BAC RP23_322B2 through homologous recombination and gap repair. The final structure of the construction is depicted in Supplementary Figure 1. Linearized targeting vector was electroporated into B6 embryonic stem cells. Correctly targeted ES clones were used for blastocyst injection, from which 12 male chimeras were produced. Chimeras were mated to B6 female mice, and germline transmission was obtained from one clone. It took several generations of breeding to remove the facilitation elements (Supplementary Figure 2), including PGK-Neo selection cassette, one FRT insertion, FLP, and Sox2-Cre alleles. Eventually, the coding exon of Nrip1 was removed and the mice carried the null allele was generated (RY-Nrip1−, Supplementary Figure 1–3). The sequences of primers for genotyping are listed in Supplementary Table 1. Heterozygous (RY-Nrip1−/+) females and males were used as breeders to produce the mice with three genotypes (RY-Nrip1+/+, RY-Nrip1−/+, and RY-Nrip1−/−) for the following experiments.
Life-Span Study
Mice in the life-span study were inspected at least once daily to collect data for time and likely cause of death. Moribund mice, which are severely ill and judged likely not to survive another 48 hours, were euthanized. Criteria for moribundity have been described previously (1). The age at which a moribund mouse was euthanized was used as the best available estimate of its natural life span.
Diet Restriction
Twenty B6 females (2 months old) were randomly divided into four cages (two control and two diet-restricted cages, five mice/cage). One control and one diet restricted cage were used for a short-term (1 month) diet restriction; the other two cages were used for long-term (4 months) diet restriction. Mice in the diet restriction group were fed a measured amount of food each day. The food quantity was 60% of the average amount eaten by control (ad-lib-fed) mice. Mice were fed at 4:00 pm each day.
Tissue Collection
Three groups of mice were euthanized for tissue collection. Group 1: for detecting the alterations of Nrip1 expression in different tissues with aging, B6 females and males at young, middle, and old ages (6, 12, and 18 months, respectively, four to seven mice/age/sex/time point); Group 2: for detecting the alterations of Nrip1 expression in female mice after diet restriction (described above, n = 5); Group 3: for investigating cellular senescence and autophagy in white adipose tissue (WAT; 18-to 22-month-old RY-Nrip1 females, four to eight mice/genotype). Following euthanization, heart, liver, kidney, small intestine, gastrocnemius skeletal muscle, ovary or prostate, and fat depots (including interscapular brown fat, inguinal, retroperitoneal, and mesenteric depots) were collected, with the additional collection of periovarian fat depots for Group 3.
Mouse Embryonic Fibroblasts
Breeder cages were set up using RY-Nrip1−/+ females and males, and the females were checked for vaginal plugs every morning. Once the vaginal plug was found, female mice were separated from males and this was considered as Day 0. At Day 14, female mice were euthanized and the embryos were collected for generating mouse embryonic fibroblasts (MEFs). Briefly, embryos were washed two times with phosphate-buffered saline, and the head and organs of the embryos were removed and saved for genotyping. Embryo sex was determined by using PCR to detect the existence of the Y chromosome-specific gene, sex determining region Y (Sry). Only females were used in this study. Primers for genotyping are listed in Supplementary Table 1.
Embryos were then minced in 60-mm cell culture dishes containing 2 mL of trypsin/ethylenediaminetetraacetic acid solution. Plates were placed in a 37°C incubator for 30 minutes. Trypsin activity was stopped by 4 mL of complete DMEM culture medium (Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum and supplement of 100 U/mL penicillin and 100 µg/mL streptomycin, Corning, 10-017-cv). Tissues were disrupted by repeated pipetting, and the suspension was transferred to 10-cm cell culture plates and allowed to grow to confluency. Media was replaced every 2 days and cells were split at 80%–90% confluency.
Amino Acid Starvation Induced Autophagy in MEF
RY-Nrip1+/+ (Wild), RY-Nrip1−/+ (Het), and RY-Nrip1−/− (Hom) MEF cell lines were tested for the effects of amino acid starvation on autophagy. Cells were seeded in six-well plates at a concentration of 2 × 105 cells/well. For the control group, cells were cultured in a complete DMEM culture medium. To induce the autophagy, cells were cultured with an amino acid-free medium, Earle’s balanced salt solution (EBSS; Sigma, E6267), with or without chloroquine (100 µM), for 1 hour. Cells were then lysed with M-PER mammalian protein extraction reagent (Thermos Scientific, 78501) to collect protein for Western blot.
Western Blot
Protein was separated by sodium dodecyl–sulfate polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes (NEN Life Science Products). Primary antibodies of LC3 (Cell Signaling Technology, #4108), NRIP1 (Santa Cluz, SC-8997), and internal control GAPDH (Cell Signaling Technology, #2118) were incubated at 4°C overnight. Primary antibodies were detected with RDye 800CW goat anti-rabbit IgG (Li-Cor, Lincoln, NE; Cat#: P/N 926–32211) by Odyssey Infrared Imaging System (Li-Cor); or goat anti-rabbit IgG, H & L chain-specific peroxidase conjugate (Millipore Sigma, #401315), using chemiluminescent substrate (Thermo Scientific, 34080).
Cell Viability Assay
MEF cell lines were seeded in six-well plates at a concentration of 3 × 104 cells/well. Media was replaced every 2 days. The rate of cellular proliferation was measured at 24, 48, and 72 hours after cell seeded. At the end of each time point, 10 µL of 5 mg/mL 3-(4,5)-dimethylthiazol-2-yl-2,5-diphenyltetrazolium bromide (MTT; Sigma, St. Louis, MO) was added to each well. Four hours later, 100 µL of dimethyl sulfoxide was added to the MTT-treated wells and the absorption at 540 nm was determined by spectrophotometry (Bio Tek PowerWave XS, Winooski, VT).
Senescence-Associated-Beta-Gal Staining
Senescent cells were detected by using Senescence Associated-Beta-Gal (SA-β-gal) kit (Cell Signal, 9860). Tissue or cells were fixed with 1× fixative buffer (provided with the kit) for 20 minutes and then stained overnight with X-gal (1 mg/mL) between 5.9 and 6.1 pH at 37°C. Staining buffer was then removed, and samples were washed with phosphate-buffered saline twice. For tissues, samples were paraffin embedded, slides were cut, and then H&E staining was performed. For cells, culture dishes were covered with 70% glycerol and stored at 4°C. To quantitate the beta-gal positive cells, for each sample, at least nine vision fields under microscope (20×, Olympus IX71) were randomly selected. In each vision field, the total number of cells and the number of positive cells were counted, and the percentage of positive cells was calculated.
Replicative and Oxidative Stress-Induced Cell Senescence
For detecting replicative senescence, MEF cell lines were seeded in six-well plates at a concentration of 5–10 × 103 cells/well. At Days 3, 5, and 7, cells were subjected to the SA-β-gal staining. To detect oxidative stress-induced senescence, MEF cell lines were seeded in six-well plates at a concentration of 3 × 104 cells/well. After 24 hours, cells were treated with H2O2 at 10 and 20 µM for 2 hours. After being washed twice with phosphate-buffered saline, cells were cultured in complete medium for another 24 hours and then subjected to SA-β-gal staining. Percentage of senescent cells was calculated as described above.
Real-Time Quantitative PCR
Total RNA was isolated by TRIzol Reagent (Invitrogen, Carlsbad, CA) using the manufacturer’s single-step chloroform-extraction protocol. Complementary DNA was synthesized using the Thermo Scientific Verso cDNA synthesis kit (Thermo Scientific, USA). An aliquot of 1 µg of total RNA was reverse-transcribed by reverse transcriptase using anchored oligo dT primers following the manufacturer’s instructions. Real-time PCR was performed using the SensiFAST SYBR Hi-ROX Kit (Bioline USA Inc.) on the ABI StepOnePlus Real-Time PCR machine (Applied Biosystems, Foster City, CA). Relative expressions of target genes were calculated with the 2−(ΔΔCT) method, using GAPDH as the reference gene. Sequences of primers are listed in Supplementary Table 2.
Statistical Analyses
Data are presented as means ± SE. Analysis of variance (ANOVA) and multiple comparisons (post hoc Tukey’s test) or Student’s t test were used to assess expression differences between groups. Survival curves were drawn using the Kaplan–Meier method. The significance of the differences among curves was determined by the log-rank test. The p values of the pairwise log-rank tests were adjusted by Bonferroni correction. p Values less than .05 were considered statistically significant. All statistical analyses were conducted by using JMP 10 (SAS Ins., Cary, NC).
Results
Knockout of Nrip1 Extends Longevity in Female Mice
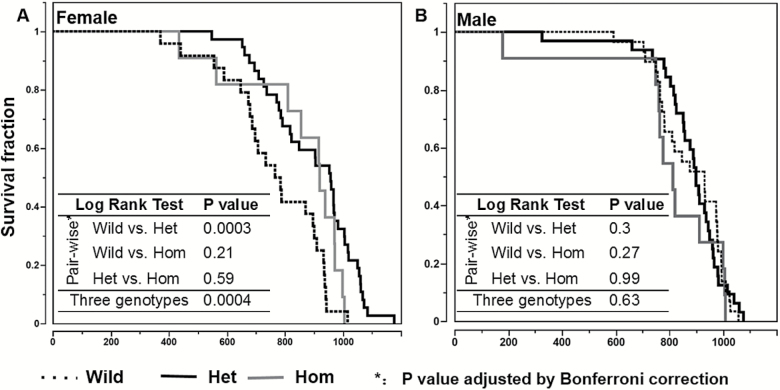
Our previous study identified five haplotypes of Nrip1 loci among the inbred strains. The haplotype that associates with delayed age of vagina patency and reduced IGF1 is also associated with extended longevity (2). We also reported delayed FSM and reduced IGF1 in the MP-Nrip1 knockout mice (2). In the current study, we found that in females, both MP-Nrip1−/+ (MP-Het) and MP-Nrip1−/− (MP-Hom) KO mice have greater longevity than wild mice. The mean life span was extended 17% and 11%, and the median life span was extended 23% and 19% compared to the MP-Wild (MP-Nrip+/+) females, respectively (Table 1). Log-rank test shows that there is a significant difference in longevity among the three genotypes of the females (Figure 1; p = .0004). Pairwise log-rank tests show that longevity in the MP-Het females is significantly greater than the longevity of MP-Wild females (Bonferroni adjusted p = .0003); however, no significant difference was found by the log-rank test between MP-Wild and MP-Hom or between MP-Het and MP-Hom. It should be noted that the Nrip1-deficient (combining the MP-Het and MP-Hom) females show significantly increased longevity compared to the MP-Wild mice (log-rank test, p = .0002; figure not shown). Importantly, the mean life span of the 20% longest-lived MP-Het females is significantly longer than that of the MP-Wild females (t test, p = .0002). In males, log-rank test shows there is no significant difference among the three genotypes (p = .63). Based on life-span data that we previously collected for 31 inbred strains, we calculate that 32 mice will be needed to detect a 10% increase in life span with a power of 0.8 (1). The relatively small sample size of the MP-Hom mice in both female and male groups may limit the statistical power. It should be noted that we used all the pups that were successfully weaned by eight breeding pairs (MP-Het × MP-Het) in 6 months for the longevity study. Based on the standard distribution of the genotypes, MP-Hom and MP-Wild mice should have similar numbers. However, among the 144 mice, only 22 are MP-Hom, which is significantly less than the MP-Wild mice (chi-square test, p < .01), suggesting that MP-Hom mice may a have higher pre-/post-natal mortality rate. Original record of life-span study can be found in Supplementary Table 3.
Deletion of Nrip1 regulates longevity differently in female and male mice. The figure shows the survival curves of female (A) and male (B) MP-Nrip1 mice. The sample sizes of the different groups are listed in Table 1. The embedded table shows the results of log-rank tests of pairwise comparisons and among three genotypes. The p values of log-rank tests are adjusted by Bonferroni correction.
Supplementary Figure 4 shows the body weight of MP-Nrip1 mice from 30 through 900 days (n ≥ 5/sex/genotype/time point). In females, both MP-Hom and MP-Het mice have significantly reduced body weight compared to the MP-Wild mice until the age of 600 days (Tukey’s test, p < .05), but no significant difference at 750 and 900 days among the three genotypes. Notably, the body weight of the MP-Hom females is about 80% of the MP-Wild females at 30 days, and this ratio drops to 70% at the age of 360 days. The differences are narrowed with aging. At 900 days, the body weights are similar, 24.2 ± 0.6, 24.3 ± 0.7, and 25.01 ± 0.8 (gram; mean ± S.E.) for MP-Hom, MP-Het and MP-Wild females, respectively. Interestingly, although the MP-Het females are significantly heavier than the Hom females in the first year, at the older ages, there is no significant difference between these two groups. In males, the MP-Wild mice are significantly heavier than the MP-Hom mice at all time points. The most significant difference is found at the age of 240 days, at which the MP-Hom males weigh 24% less than MP-Wild mice (23.8 ± 0.9 vs. 31.2 ± 0.8 g). Between 240 and 600 days, the MP-Wild males are also significantly heavier than the MP-Het males. Although the MP-Het males are heavier than the MP-Hom males at all time points, only at 120 and 240 days the differences are significant.
Nrip1 Expression Is Altered Differently by Aging in Different Tissues and Sexes
To further investigate the role of Nrip1 in aging, we measured the mRNA expression levels of Nrip1 in 11 different tissues at young (6 months), middle (12 months), and old (18 months) ages for both sexes (four–seven mice/sex/age) using real-time quantitative PCR (RT-qPCR). A two-way ANOVA was conducted that examined the effect of sex and age on the expression of Nrip1 for each tissue, except for the ovary and prostate, in which only age was included in the model. Except for inguinal white fat, ovary and prostate, a significant difference in expression was detected in all other tissues (Table 2). There was a statistically significant interaction between sex and age on expression of Nrp1 in brown fat (p = .005), heart (p = .019), and liver (p = .021).
Table 2.
Summary of the p Values of the Comparisons of Nrip1 Expression in Different Tissues at Different Ages of Female and Male Mice
| ANOVA p Value | Post Hoc Tukey’s Test p Value | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sex | Y vs. M | Y vs. O | M vs. O | F vs. M at | |||||||
| Whole Model | Sex | Age | Sex × Age | Y | M | O | |||||
| Bft | <.001 | <.001 | .002 | .005 | F | 1.000 | 1.000 | 1.000 | .015 | .001 | 1.0 |
| M | .997 | .007 | <.001 | ||||||||
| Gas | <.001 | <.001 | .687 | .413 | F | 1.000 | .997 | .998 | .001 | <.001 | <.001 |
| M | .765 | .735 | 1.000 | ||||||||
| Hrt | <.001 | <.001 | .06 | .019 | F | .064 | .011 | .996 | <.001 | .022 | .088 |
| M | 1.000 | .999 | 1.000 | ||||||||
| Ing | .659 | .453 | .656 | .428 | F | 1.000 | 1.000 | 1.000 | .935 | .929 | .986 |
| M | 1.000 | .715 | .750 | ||||||||
| Kid | <.001 | <.001 | .075 | .134 | F | .999 | 1.000 | 1.000 | <.001 | <.001 | <.001 |
| M | .623 | .046 | .545 | ||||||||
| Liv | .014 | .031 | .093 | .021 | F | .991 | .973 | 1.000 | 1.000 | .998 | .009 |
| M | .998 | .044 | .046 | ||||||||
| Mes | .004 | <.001 | .227 | .727 | F | 1.000 | .871 | .923 | .311 | .037 | .199 |
| M | .785 | .737 | 1.000 | ||||||||
| Ret | <.001 | <.001 | .019 | .051 | F | .999 | .022 | .035 | .351 | .039 | <.001 |
| M | .995 | 1.000 | .993 | ||||||||
| Sl | <.001 | <.001 | .139 | .186 | F | 1.000 | 1.000 | .999 | <.001 | <.001 | <.001 |
| M | .151 | .122 | .999 | ||||||||
| Ova | .536 | Not apply | F | .840 | .858 | .506 | Not apply | ||||
| Pro | .49 | M | .736 | .889 | .473 | ||||||
Note: M = middle; O = old; Y = young. Numbers in bold are less than .05.
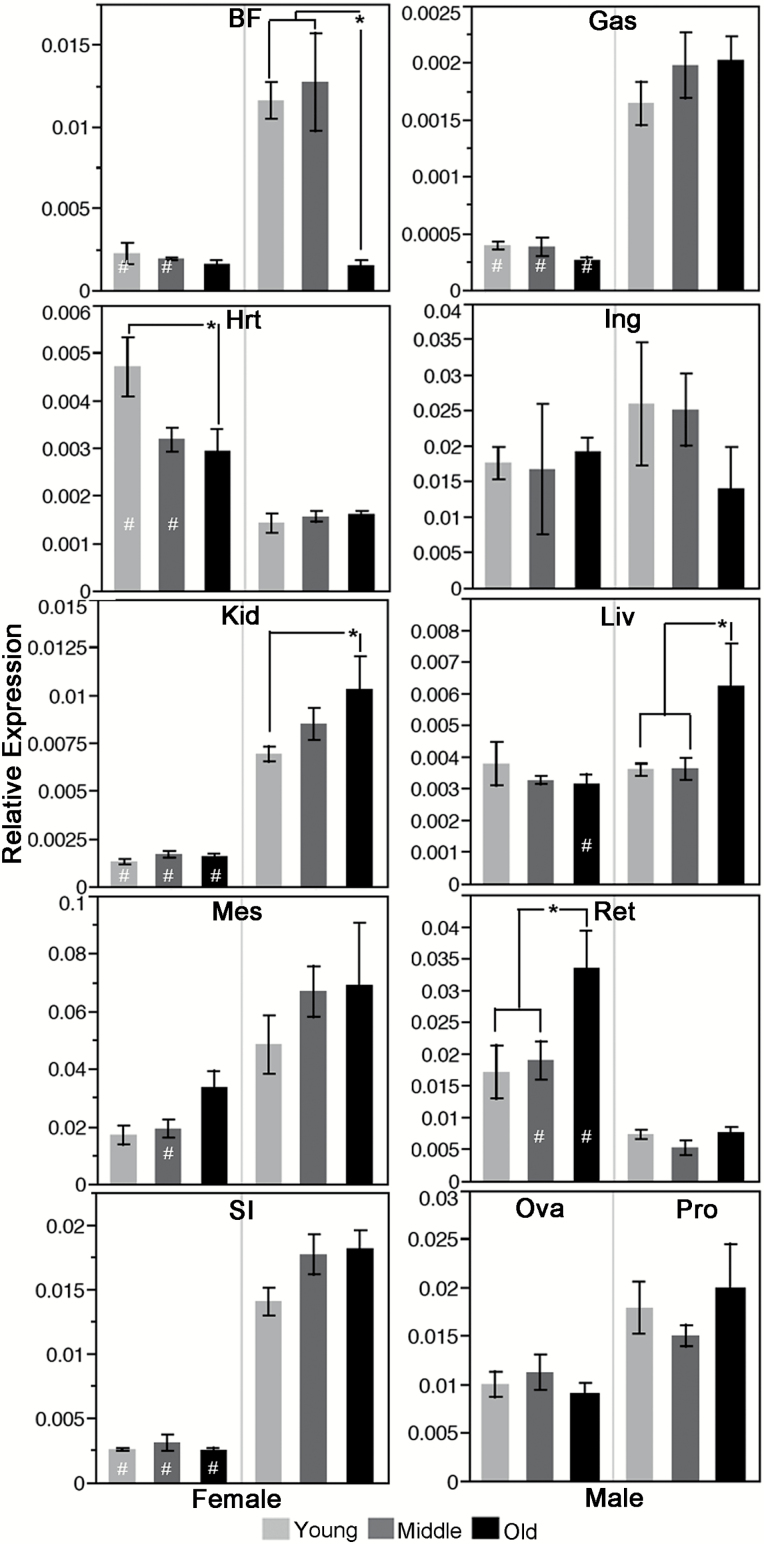
When comparing Nrip1 expression between males and females at each of the three time points, female mice had significantly lower expression levels in the gastrocnemius muscle, kidney, and small intestine at all three time points (Table 2; Figure 2). In brown fat, female mice have significantly lower expression of Nrip1 than male mice at both young and middle ages (p = .015 and p = .001, respectively). In the liver, females have a significantly lower expression of Nrip1 compared to males at old age (p = .009). However, in both the heart and retroperitoneal WAT, at two of the three time points, female mice have significantly higher expression than male mice. No significant difference was found between males and females at any of the time points in inguinal white fat tissue.
Nrip1 expression is altered differently among tissues/organs and sexes during aging. Using RT-qPCR, Nrip1 expression in different tissues/organs at different ages (6, 12, and 18 months) was measured for both females and males (4 ≤ n ≤ 7). *Comparison within one sex at different ages, Tukey’s test p < .05. #Comparison between sexes at the same age, Tukey’s test p < .05. Bft = brown fat; Ing = inguinal white adipose tissue; Mes = mesenteric white adipose tissue; Ret = retroperitoneal adipose tissue; Gas = gastrocnemius; Hrt = heart; Kid = kidney; Liv = liver; SI = small intestine; Ova = ovary; Pro = prostate.
In females, Nrip1 expression in retroperitoneal WAT is significantly upregulated at old age compared to its levels at young and middle ages (p = .022, p = .035, respectively). In the heart, Nrip1 expression is significantly downregulated at old age versus its expression at young age (p = .011). In males, the most significant change was found in brown fat at old age, in which the Nrip1 expression is approximately six times lower than its expression at young and middle ages, whereas in kidney and liver, Nrip1 expression is significantly upregulated at old age compared to young age (Table 2; Figure 2).
ANOVA and post hoc Tukey’s analyses also showed that there are significant differences in Nrip1 expression among tissues/organs at each time point (p < .0001). Interestingly, WAT (inguinal, retroperitoneal, and mesenteric) in female mice always has the highest expression level of Nrip1, whereas the gastrocnemius skeletal muscle always has the lowest expression (Supplementary Table 4). In male mice, mesenteric WAT has a significantly higher Nrip1 expression level than all other tissues at all three time points (Tukey’s test, p < .05; Supplementary Table 4).
Nrip1 Expression Is Altered Differently in Different Tissues in Response to Diet Restriction
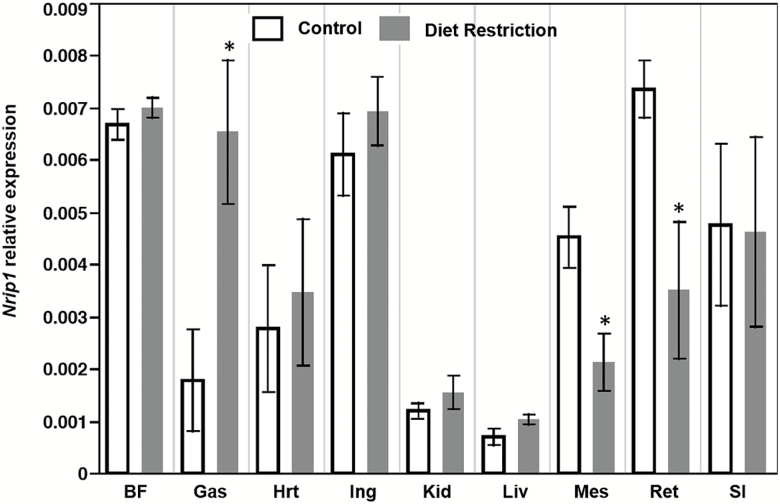
In the 1-month diet restriction study, 40% food reduction did not induce significant alteration in Nrip1 expression among the nine tested tissues/organs (data not shown). In the four-month restriction study, t test shows expression of Nrip1 was significantly reduced in mesenteric and retroperitoneal WAT (n = 5, p < .05; Figure 3), but significantly upregulated in the gastrocnemius muscle. No significant differences were found in other tissues. These results indicate that that Nrip1 expression varies in different tissues in response to diet restriction.
The Deletion of Nrip1 Elevates the Autophagy in MEFs
To investigate the role of Nrip1 in aging at the cellular level, we created MEF cell lines from RY-Nrip1+/+ (Wild), RY-Nrip1−/+ (Het), and RY-Nrip1−/− (Hom) female embryos. The representative genotyping results of the RY-Nrip1 alleles and Y chromosome gene, Sry, are shown in Supplementary Figures 3 and 5D, respectively. The deletion of Nrip1 was also verified by RT-qPCR and Western blot (Supplementary Figure 5A–C). The RY-Wild MEF has a significantly higher expression of Nrip1 at both mRNA and protein levels than the RY-Het MEF. In the RY-Hom MEF, the mRNA and protein are not detectable.
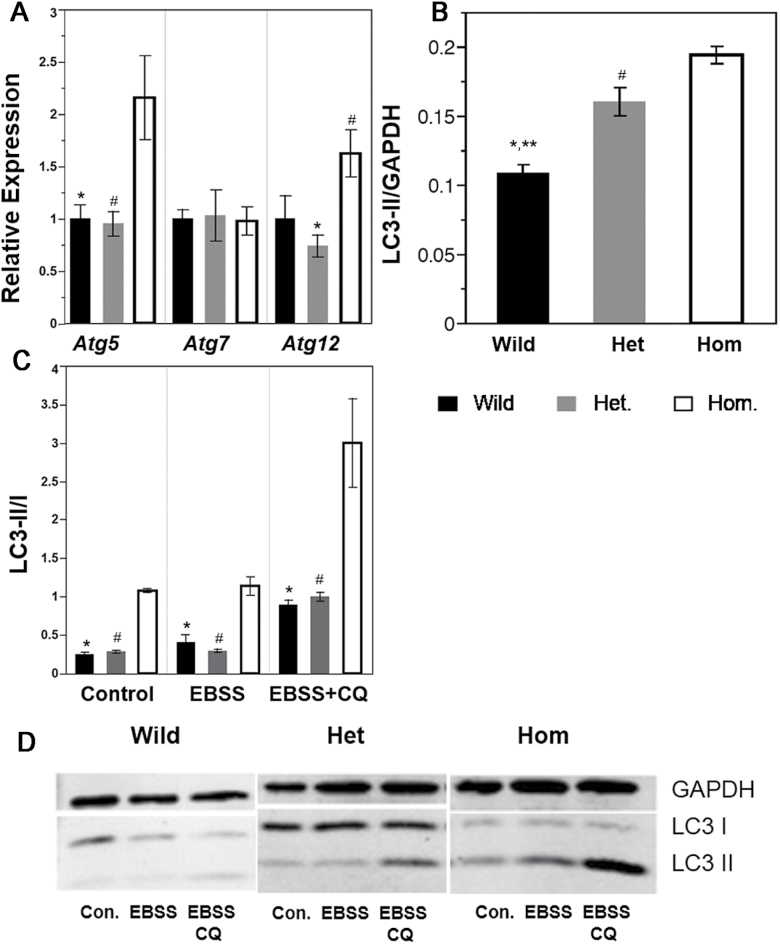
Because autophagy has been identified as a key cellular mechanism that profoundly impacts aging, we tested for the involvement of Nrip1 in regulating cell autophagy. Interestingly, compared to the RY-Het and RY-Wild MEF, the homozygous knockout of Nrip1 significantly upregulated the autophagy-related genes (ATG) Atg5 and Atg12, but not Atg7 (Figure 4A; Tukey’s test, p < .05, n ≥ 4), thus strongly suggesting that autophagy may be altered.
Depletion of Nrip1 in embryonic fibroblasts elevates autophagy. (A) The expression of autophagy-related genes, Atg5, Atg7, and Atg12, were tested by RT-qPCR (n ≥ 4). MEFs were subjected to the starvation study; LC3 were detected by Western blot (n ≥ 3). (B) Under the control condition, the autophagy status is shown by LC3-II/GAPDH. (C) The autophagy flux of control, Earle’s balanced salt solution (EBSS) amino acid starvation, and EBSS with chloroquine (CQ) 1-hour treatment is shown by LC3-II/I. (D) Representative images of the Western blot. GAPDH were used as loading control. *Wild versus Hom, **Wild versus Het, #Het versus Hom, Tukey’s test p < .05.
Autophagosome engulfs cytoplasmic components during autophagy, causing the cytosolic form of LC3 (LC3-I) to be conjugated to phosphatidylethanolamine to form LC3-phosphatidylethanolamine conjugate (LC3-II), which is recruited to the autophagosomal membrane. LC3-II is then degraded after fusion of the autophagosome with the lysosome (autolysosome). Thus, the autophagosomal marker LC3-II can reflect autophagy activity. Autophagy inhibitor chloroquine can inhibit the fusion of autophagosome and lysosome, and in this way, prevents the digestion of LC3-II. Thus, in the chloroquine-treated cells, the ratio between LC3-II and LC3-I can reflect the autophagy flux. To understand the effect of the depletion of NRIP1 on autophagy, we first examined autophagy activity under the normal culture condition. As shown in Figure 4B, the ratio between LC3-II and GAPDH is significantly elevated in the Het and Hom MEFs, compared to the Wild MEF. The Hom MEF also has a significantly higher ratio than that of the Het MEF (Figure 4B and andD;D; Tukey’s test p < .05, n ≥ 3). These results indicate that the deletion of Nrip1 upregulates the basal level of autophagy. Next, we examined autophagy flux in the MEF cell lines under control, EBSS amino acid starvation with or without chloroquine. There was no significant difference in the LC3-II/I ratio between Het and wild-type MEFs. However, Hom MEFs have a significantly elevated LC3-II/I ratio than that of Het and Wild MEFs under all three conditions, indicating that the homozygous knockout of Nrip1 can upregulate autophagy flux (Figure 4C and andD;D; Tukey’s test p < .05).
The Deletion of Nrip1 Suppresses Proliferation and Delays Senescence in MEFs
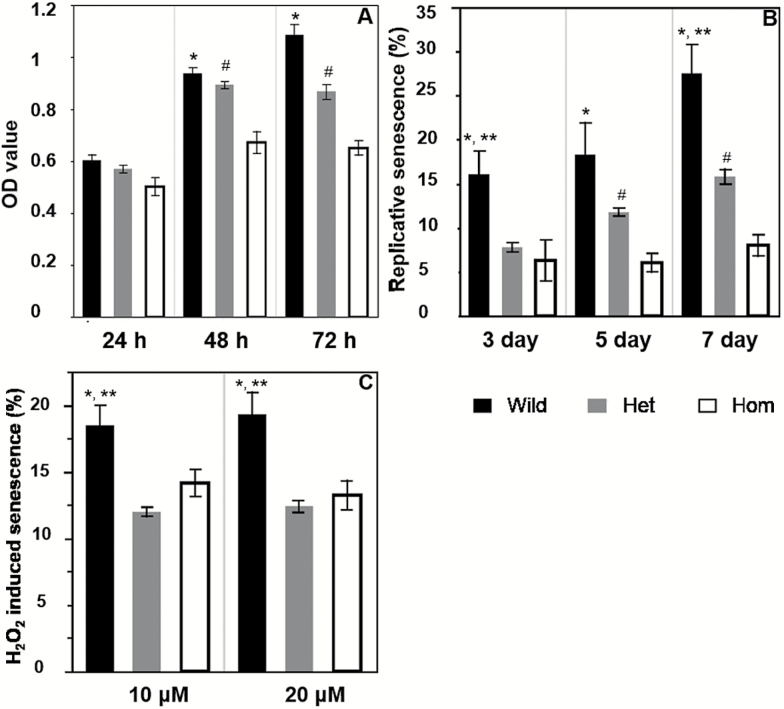
MTT assay shows that at 48 and 72 hours, the Hom MEF has significantly reduced proliferation than the Het and Wild MEF (Figure 5A; Tukey’s test, p < .05). At all three time points, although the OD values of Het MEFs are lower than that of Wild MEFs, the differences are not significant.
Depletion of Nrip1 in embryonic fibroblasts reduces proliferation and delays cellular senescence. (A) Cell viability was tested by the MTT method; (B) replicative senescence was examined at Days 3, 5, and 7. (C) Oxidative stress-induced senescence was examined by treating cells with 10 and 20 µM H2O2. *Wild versus Hom, **Wild versus Het, #Het versus Hom, Tukey’s test, p < .05, n ≥ 5.
We further compared the proliferative senescence (Figure 5B) and oxidative stress-induced senescence (Figure 5C) detected by SA-β-gal staining among the three genotypes of the female MEFs (n ≥ 5). We found that the cell numbers of Wild MEFs were significantly reduced after Day 7 and when the H2O2 concentration was higher than 40 µM, suggesting the cell death is triggered under these conditions (data not shown). Between Day 3 and Day 7, the percentage of senescent cells increased 72% (16.0 ± 2.7% vs. 27.5 ± 3.2%) in Wild MEFs, 102% in Het MEFs (7.8 ± 0.5% vs. 15.8 ± 0.8%), and 27% in Hom MEFs (6.3 ± 2.3% vs.8.0 ± 1.2%). At all three time points, Hom MEF had significant fewer senescent cells than Wild MEFs. At Days 3 and 7, the Het MEFs also has a significantly lower percentage of senescent cells than the Wild MEFs; and, at Days 5 and 7, Hom MEFs had significantly lower percentages of senescent cells than the Het MEFs (Tukey’s test, p < .05). Representative photographs of senescent cells on Day 7 are shown in Supplementary Figure 6. These results strongly suggest that replicative senescence is delayed by the depletion of NRIP1. Oxidative stress-induced senescence was also suppressed by the deletion of NRIP1. As shown in Figure 5C, there were significantly fewer senescent cells in the Hom and Het MEFs compared to the Wild MEFs after 10 and 20 µM H2O2 treatments. No significant difference in the H2O2 induced senescence was found between the Het and Hom MEFs.
The Deletion of Nrip1 Reduces Cellular Senescence in WAT
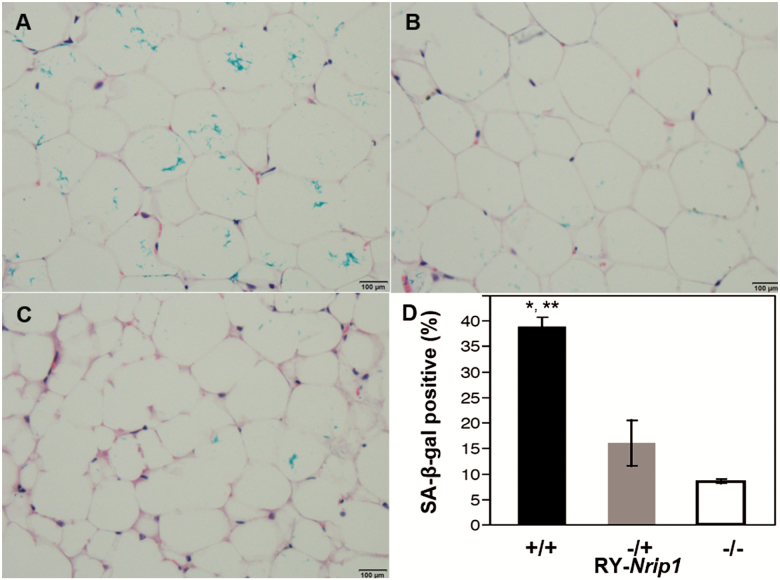
The periovarian WAT (EWAT) forms of RY-Nrip1+/+, RY-Nrip1−/+, and RY-Nrip1−/− old females (18–22 months) were stained with SA-β-gal. The tissues were then paraffin embedded for histology study. As shown in Figure 7, compared to the RY-Nrip1+/+, EWAT of RY-Nrip1−/+ and RY-Nrip1−/− have significantly fewer senescent cells (Figure 6; n ≥ 3, Tukey’s test, p < .05).
Depletion of Nrip1 in mice delays cellular senescence in white adipose tissue. At old age (n ≥ 3, 18–22 months), senescent cells in periovarian fat depot of the wild-type (A), RY-Nrip1−/+ (B), and RY-Nrip1−/− (C) females were detected by SA-β-gal staining. Statistical analysis detected significant difference (D). *Wild versus Hom, **Wild versus Het, Tukey’s test, p < .05.
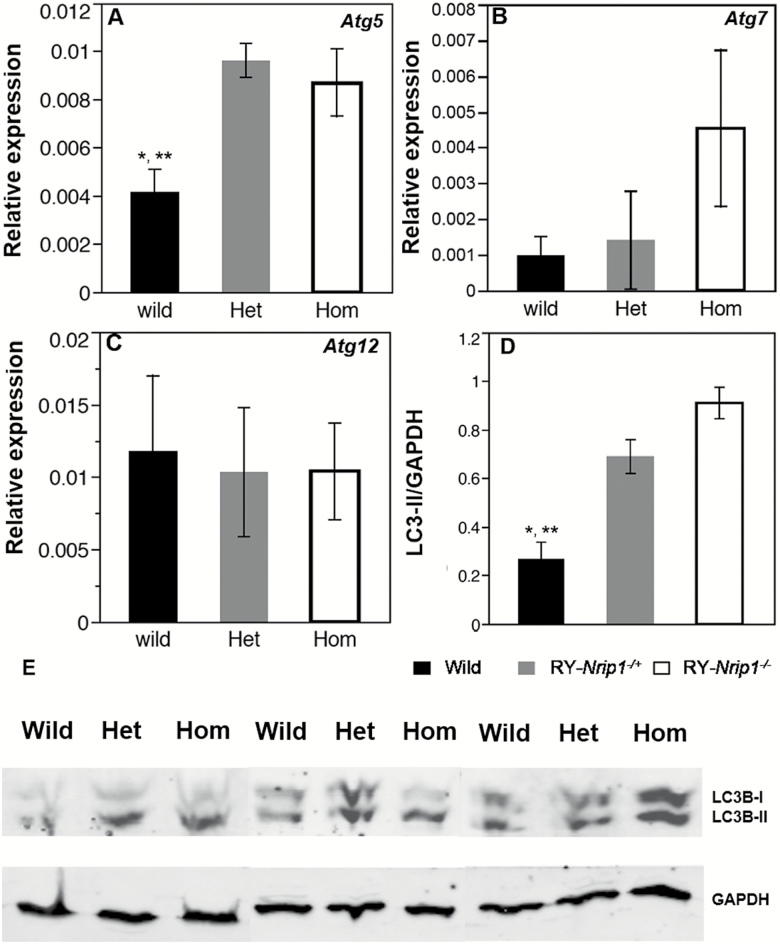
Depletion of Nrip1 in mice elevates autophagy in periovarian white adipose tissue at old age. Autophagy-related genes 5, 7, and 12 were measured by RT-qPCR (A–C). The autophagy status (LC3-II/GAPDH) was examined by Western blot (D, E). *Wild versus Hom, **Wild versus Het, Tukey’s test, p < .05, n ≥ 3.
Because the increased autophagy status has been found in MEFs, and autophagy may play an important role in regulating cell senescence (13), and the autophagy is elevated in Nrip1-depleted MEF, we examined the possibility of altering autophagy by the depletion of NRIP1 in WAT. RT-qPCR shows that Atg5 has significantly higher expression in RY-Nrip1−/+ and RY-Nrip1−/− EWAT than in RY-Nrip1+/+ EWAT (Figure 7A; Tukey’s test, p < .05, n ≥ 3), but no significant difference was found for the expressions of Atg7 and Atg12 (Figure 7B and andC).C). It is worth noting that Atg7 expression is four times higher in the RY-Nrip1−/− EWAT than that in RY-Nrip1+/+ EWAT, although it is not statistically significant (Tukey’s test, p = .12). The elevated autophagy in EWAT by the depletion of NRIP1 is verified by detecting LC3 by Western blot. As shown in Figure 7D and andE,E, the ratio of LC3-II and GAPDH, which reflects the basal level of autophagy, is significantly elevated in both RY-Nrip1−/+ and RY-Nrip1−/− EWAT (n = 3, Tukey’s test, p < .05).
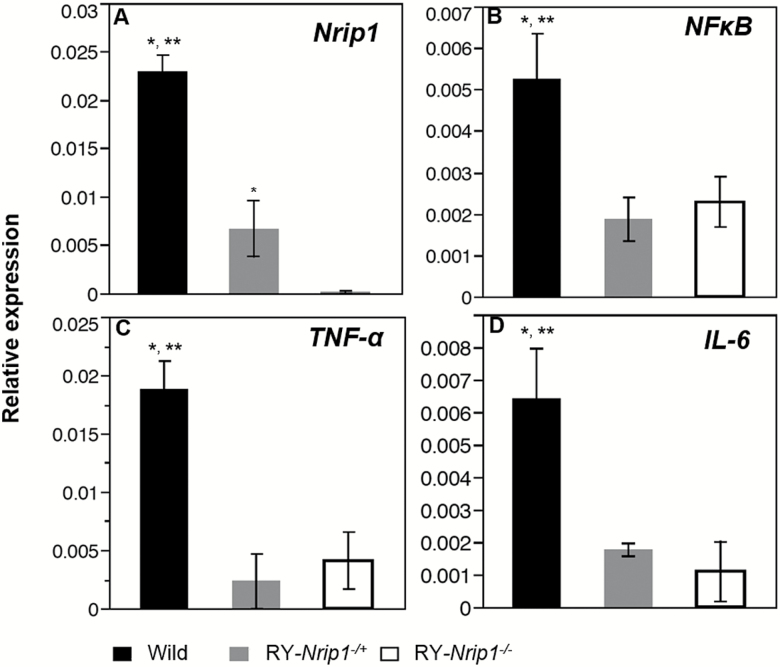
The senescent adipocyte is one of the important resources of proinflammatory cytokines. RT-qPCR found that along with the significantly reduced senescence in EWAT, the expression of nuclear factor-κB (NF-κB), tumor necrosis factor-α (TNF-α), and interleukin-6 (IL-6) is all significantly suppressed (Figure 8; n ≥ 3, Tukey’s test, p < .05).
Discussion
In the past 30 years, by combining gene mapping, such as quantitative trait loci and bioinformatics analyses, many disease-related genes have been identified (14–16). Using longevity as a biomarker of aging, gerontologists have conducted a series of genetic mapping studies in mice and tried to identify the core aging genes (reviewed in ref. (17)). Although genetic loci that associate with longevity have been reported, no aging or longevity gene has been identified by using this methodology. Two major reasons are responsible for this frustration. First, longevity is one of the most complicated biological traits, which can be affected by innumerable genetic and environmental causes and their interactive factors. Therefore, each longevity-related regulatory locus can only explain a small amount of the variation in life span, making the identification difficult. Second, using longevity as a biomarker for a mammalian aging study is extremely time consuming. Gerontologists have searched for a biomarker that shares common genetic and molecular regulatory mechanisms with longevity and that could be measured at young age but accurately predict longevity. Our previous studies found that among mouse inbred strains, the circulating IGF1 level and the age of FSM associated with longevity. Using these two biomarkers, we have identified a group of candidate genes (1,2,18–20). Among these genes, we verified that the knockout of Nrip1 can delay FSM and reduce circulating IGF1 levels (2).
Global Suppression of Nrip1 Extends Female Longevity
In the current study, our results clearly show that the deletion of Nrip1 in females can significantly extend longevity compared to wild-type females, although the difference between the MP-Nrip1−/− and MP-Nrip1+/+ females is not statistically significant. The median life span of the MP-Nrip1−/+ females is extended by 23% (958 vs. 776 days), and the mean life span of the longest 20% lived females is extended by 14% (1083 vs. 954.5 days, p = .0002) compared to the MP-Nrip1+/+ females. Although the extension of longevity was not found in the Nrip1 knockout males, the positive effects of Nrip1 knockout on metabolism are not sex specific (10,21,22). The difference in female and male longevity has been observed in many animal models (reviewed in ref. (17)) and in human populations (23). However, the underlying genetic and molecular mechanisms remain vague. Given the fact that NRIP1 has profound interactions with nuclear receptors (21,22,24), including estrogen receptor and androgen receptor, it is reasonable to hypothesize that the NRIP1 suppression may have different biological or pathological effects on aging-related diseases and frailties in different sexes.
White Adipose Tissue Might Be a Proper Target for Tissue-Specific Manipulation of Nrip1 to Extend Longevity
Although the global knockout (GKO) of Nrip1 can improve metabolism and female longevity, serious detrimental effects have been reported. Nrip1 expression in ovaries is essential for reproduction. GKO of Nrip1 causes female infertility (8) along with memory impairment (25) and developmental retardation (7). Importantly, emerging studies of cancers have revealed that NRIP1 has antagonistic effects in tumorigenesis. For instance, the suppression of NRIP1 accelerates the growth of colon epithelial cells and may promote the progression of colon cancer (21). Using transgenic mice, Lapierre and colleagues reported that the upregulation of NRIP1 suppressed colon cancer growth (21). However, our previous study found that in the mammary gland, NRIP1 depletion significantly suppressed growth of, and induce apoptosis in, the epithelial cells. Using MP-Nrip1 knockout mice, we demonstrated that the suppression of NRIP1 reduced incidence, delayed onset, and suppressed progression of chemically induced breast cancer (12). The relative shorter longevity in MP-Nrip1−/− females than the longevity of MP-Nrip1−/+ females may reflect the fact that the longevity is the overall outcome of the GKO of Nrip1, which has both favorable and unfavorable effects. Taken together, these studies suggest that to maximize the positive effects on health span, Nrip1 needs to be manipulated in a tissue-specific manner.
To understand the tissue-specific role of NRIP1 in aging, it is important to examine the function of NRIP1 in different tissues at different ages. The function of NRIP1 is determined not only by the expression level but also by the post-translational modifications and subcellular distribution (26–29). In the current study, we focused on the alterations of Nrip1 expression at the mRNA level during aging and under diet restriction, which needs to be further verified by Western blot. The results show that in females, Nrip1 expression in visceral WAT is elevated at old age and that the elevation in retroperitoneal WAT is significant. Importantly, diet restriction can significantly reduce Nrip1 expression in mesenteric and retroperitoneal WAT. This expression pattern of Nrip1 in WAT may reflect decreasing metabolism in aging that can be improved by diet restriction. Given the fact that the deletion of Nrip1 in adipocytes can enhance mitochondrial function and increase the uptake of glucose (reviewed in ref. (3)), as well as the documented robust effects of diet restriction on improving metabolism, it is reasonable to hypothesize that targeting WAT only for Nrip1 deletion may improve metabolism in aging.
The importance of investigating WAT-specific manipulation of Nrip1 is further strengthened by the finding of reduced cellular senescence in WAT of RY-Nrip1-deficient mice. The important role of WAT in aging has been emerging in recent years (reviewed in ref. (30)). WAT is not only the storage space for lipids, but it is also a major source of circulating inflammatory cytokines, such as NF-κB, TNF-α, and IL-6, which all have a strong impact on insulin sensitivity. During the aging process, accumulated senescent preadipocytes in WAT limit adipocyte proliferation, directly inducing the failure of lipid storage, thus leading to systemic lipotoxicity. The detrimental effects of senescent WAT are intensified by senescence-associated secretory phenotype (SASP) secretion, by senescent preadipocytes, and by infiltrated macrophages (30). The enhanced inflammatory profile in WAT is directly related to diabetes and obesity (31), both of which are important risk factors of reduced health span. Previous studies suggest that Nrip1 deletion in macrophages can significantly reduce the secretion of proinflammatory cytokines (27). We recently reported that in the human keratinocytes, knocking down Nrip1 by shRNA can significantly reduce the expression of NF-κB (32). Although the underlying mechanism is not clear, our current results demonstrate that the deletion of Nrip1 suppresses senescence not only in vitro in MEFs but also in vivo in WAT. One potential mechanism may be related to the reduced IGF1 level, which has also been suggested as a major mechanism in delaying senescent cell accumulation in WAT of long-lived GH/IGF1-deficient mice (33). Interestingly, in WAT the expression of NF-κB, as well as the proinflammatory cytokines TNF-α and IL-6 are significantly suppressed by the deletion of Nrip1. However, the inflammation profile of WAT needs to be further verified at the protein level. Furthermore, investigation is needed to test if the alteration of the inflammation profile in WAT can alter the profile of circulating inflammatory cytokines. Importantly, a recent study showed that depleting senescent cells in 18-month-old INK-ATTAC mice can enhance adipogenesis, improve WAT function, and insulin sensitivity (34). Taken together, our study suggests that WAT-specific Nrip1 deletion may be a feasible therapeutic method for reducing senescent cells in WAT and extending health span.
Nrip1 Knockout in Skeletal Muscle May Have Complex Effects on Metabolic Aging
Skeletal muscles play an important role in whole-body metabolism as they account for approximately 70% of the total insulin-stimulated glucose uptake. Type I fibers (oxidative fibers, slow twitch) and type II fibers (glycolytic fibers, fast twitch) play different roles in metabolic disorders and aging-related metabolism decline. In insulin-resistant states, such as obesity and type 2 diabetes, insulin-stimulated glucose uptake is markedly impaired (35), whereas increasing the number of type I fibers enhances insulin-mediated glucose uptake and protects against diabetes and other metabolic diseases (36). Moreover, in obese patients, skeletal muscles have reduced metabolic oxidative capacities with morphologic changes that reveal decreased type I slow-twitch fibers (reviewed in ref. (3)). However, during aging, compared to the type I fibers, type II fiber atrophy has earlier onset and more severe progress. The decline in skeletal muscle mass with aging is mainly attributed to a reduction in type II muscle fiber size (37). Akasaki used a conditional transgenic mouse model that expresses a constitutively active form of skeletal-muscle-specific Akt1 to restore glycolytic fast-twitch muscle fibers. Using this model, the authors found that restoration of glycolytic fast-twitch muscle fibers led to a reduction in fat mass and hepatic steatosis in older animals, and improved insulin sensitivity. Therefore, maintaining the type II fibers might be critical for maintaining metabolic health during aging.
A previous study found that Nrip1 is expressed in a fiber type-specific manner in skeletal muscle. At the mRNA level, it is expressed at a higher level in type II glycolytic muscles like gastrocnemius and extensor digitorum longus (EDL) muscle compared with the type I oxidative muscles like soleus (38). Using Nrip1 global knockout mice and transgenic mice that over-express Nrip1, previous studies revealed that the Nrip1 expression level regulates the proportions of type I and type II fibers in muscle. In NRIP1 null mice, mitochondrial activity is increased in muscle fibers, the glycolytic muscles are redder and contain more type I than the wild-type mice. Gene expression involved in fatty acid oxidation, oxidative phosphorylation, and mitochondria biogenesis is upregulated in the type II (the glycolytic) muscle in NRIP1 null mice. Importantly, oxidative muscles have no detectable change in phenotype, as they are predominantly type I fiber and express a low level of NRIP1 (38). The elevation of NRIP1 expression results in an increased number of type II fibers and in decreased mitochondria activity (38). In the current study, we found that Nrip1 expression in the gastrocnemius, the glycolytic muscle, was upregulated significantly after 4 months of calorie restriction. This may indicate a decreased proportion of type I fiber, which is explainable by the reduced fatty acid resource utilized by type I fiber. Therefore, the deletion of Nrip1 in skeletal muscle may have important effects on metabolic dysfunction during aging. Given the fact that diet restriction has robust effects on improving metabolism, but increases the expression of Nrip1 in the gastrocnemius, it is not unreasonable to hypothesize that deletion of Nrip1 in skeletal muscle may have negative effects on metabolic aging and health span.
Enhanced Autophagy by Nrip1 Deletion May be a Critical Mechanism for Improving Metabolic Aging
Our in vitro studies suggest that under normal and amino acid starvation conditions, the deletion of Nrip1 may enhance autophagy. Autophagy is an evolutionarily conserved process for cellular homeostasis through the degradation of long-lived proteins and functionally redundant or damaged intracellular organelles in lysosomes. It has also been reported that autophagy plays an important role in the regulation of inflammation through several different mechanisms that have not been thoroughly clarified. It has been suggested that in dendritic cells, autophagy induction improves antigen presentation (39), and several major immune effectors (including T lymphocytes) rely on autophagy for the maintenance of their function (40). Disabled autophagy provokes the accumulation of p62/STQM1, which activates the proinflammatory transcription factor NF-κB, and the stress-responsive transcription factor NRF2, thereby favoring inflammation and tissue injury (40). Our results show that the deletion of Nrip1 in MEF and in WAT of old animals can enhance autophagy significantly. This might be one of the underlying mechanisms that suppress the expressions of Nfkb1 and proinflammatory cytokines, IL-6 and TNF-α. Autophagy has also been suggested in the efficiency of dead cells clearance by apoptosis. It has also been suggested that insufficient autophagy could stimulate inflammatory responses secondary to reduced clearance of dead cells (41). Therefore, the upregulated autophagy may also suppress proinflammation by facilitating apoptosis. Indeed, we have reported that suppressing Nrip1 in epithelial cells and keratinocytes can significantly induce apoptosis in vitro (12,32).
Notably, in regard to WAT, there are dramatic differences between sexes, including its body location, accumulation, secretory profile of cytokines, reaction to hormones, and effect on metabolic rate (42–44). Regarding autophagy, it has also been reported that constitutive autophagy and stress-induced autophagy are differently regulated in several organs/tissues (45–47) between sexes. Current report focuses on the senescence and autophagy in female MEF and female WAT. Since NRIP1 has profound interactions with estrogen receptor and androgen receptor (7,48), it will be interesting to test if suppressing NRIP1 has different biological or pathological effects on WAT function in aging in different sexes.
It is worth noting that enhanced autophagy has been considered a key mechanism for delayed aging by the suppression of insulin/IGF1/mTOR signaling (49). The reduced circulating IGF1 level in Nrip1 knockout mice (2) suggests that there might be an interaction between NRIP1 and the insulin/IGF1/mTOR pathway. Elevated autophagy by suppressing NRIP1 under normal and amino acid starvation conditions observed in this study further supports this hypothesis. Furthermore, because of the interactions of NRIP1 with estrogen receptor and androgen receptor, it will be interesting to investigate if NRIP1 plays a role in the sex difference of delaying aging by the genetic or pharmacological suppression of mTOR (23,50).
Conclusion
Our study revealed that the deletion of Nrip1 in female mice could extend longevity in addition to its effect on delaying FSM. Nrip1 expression profile may be altered differently in different tissues/organs during aging and under diet restriction. Several lines of evidence suggest that deleting Nrip1 specifically in WAT may improve metabolic aging, whereas its deletion in skeletal muscle may have an opposite effect. In vitro and in vivo studies suggest that delayed senescence and enhanced autophagy may be critical mechanisms of extending health span by deleting Nrip1.
Funding
This work is supported by NIH grants R21 AG034349, K01AG046432, and R03AG046605 to R.Y.
Supplementary Material
Supplementary data is available at The Journals of Gerontology, Series A: Biological Sciences and Medical Sciences online.
Supplement Tables
Supplement Fig1
Supplement Fig2
Supplement Fig3
Supplement Fig4
Supplement Fig5
Supplement Fig6
Legends of Supplement Figures
Acknowledgments
Lisa Hensley kindly edited the manuscript. Division of Laboratory Animal Medicine of SIUSOM provides excellent environment for animal research.