- Journal List
- Aging Cell
- v.17(4); 2018 Aug
- PMC6052393

Lifelong reduction in complex IV induces tissue‐specific metabolic effects but does not reduce lifespan or healthspan in mice
 1
,
2
,
9
1
,
2
,
9
Sathyaseelan S. Deepa
1 Aging and Metabolism Research Program, Oklahoma Medical Research Foundation, Oklahoma City, Oklahoma
Gavin Pharaoh
1 Aging and Metabolism Research Program, Oklahoma Medical Research Foundation, Oklahoma City, Oklahoma
2 Department of Physiology, University of Oklahoma Health Sciences Center, Oklahoma City, Oklahoma
Michael Kinter
1 Aging and Metabolism Research Program, Oklahoma Medical Research Foundation, Oklahoma City, Oklahoma
Vivian Diaz
3 Department of Cellular and Structural Biology, Barshop Institute for Longevity and Aging Studies, University of Texas Health Science Center San Antonio, San Antonio, Texas
Wilson C. Fok
4 Division of Hematology, Department of Medicine, Washington University in St. Louis, St. Louis, Missouri
Kaitlyn Riddle
1 Aging and Metabolism Research Program, Oklahoma Medical Research Foundation, Oklahoma City, Oklahoma
Daniel Pulliam
1 Aging and Metabolism Research Program, Oklahoma Medical Research Foundation, Oklahoma City, Oklahoma
3 Department of Cellular and Structural Biology, Barshop Institute for Longevity and Aging Studies, University of Texas Health Science Center San Antonio, San Antonio, Texas
Shauna Hill
1 Aging and Metabolism Research Program, Oklahoma Medical Research Foundation, Oklahoma City, Oklahoma
3 Department of Cellular and Structural Biology, Barshop Institute for Longevity and Aging Studies, University of Texas Health Science Center San Antonio, San Antonio, Texas
Kathleen E. Fischer
5 Biology Department, University of Alabama at Birmingham, Birmingham, Alabama
Vanessa Soto
3 Department of Cellular and Structural Biology, Barshop Institute for Longevity and Aging Studies, University of Texas Health Science Center San Antonio, San Antonio, Texas
Constantin Georgescu
6 Arthritis & Clinical Immunology Research Program, Division of Genomics and Data Sciences, Oklahoma Medical Research Foundation, Oklahoma City, Oklahoma
Jonathan D. Wren
6 Arthritis & Clinical Immunology Research Program, Division of Genomics and Data Sciences, Oklahoma Medical Research Foundation, Oklahoma City, Oklahoma
Carlo Viscomi
7 MRC‐Mitochondrial Biology Unit, University of Cambridge, Cambridge, UK
Arlan Richardson
8 Department of Geriatric Medicine, Reynolds Oklahoma Center on Aging, University of Oklahoma Health Sciences Center, Oklahoma City, Oklahoma
9 Oklahoma City VA Medical Center, Oklahoma City, Oklahoma
Holly Van Remmen
1 Aging and Metabolism Research Program, Oklahoma Medical Research Foundation, Oklahoma City, Oklahoma
2 Department of Physiology, University of Oklahoma Health Sciences Center, Oklahoma City, Oklahoma
9 Oklahoma City VA Medical Center, Oklahoma City, Oklahoma
 Corresponding author.
Corresponding author.Holly Van Remmen, Aging and Metabolism Research Program, Oklahoma Medical Research Foundation, Oklahoma City, OK.
Email: gro.frmo@nemmeRnaV-ylloH,
Associated Data
Summary
Loss of SURF1, a Complex IV assembly protein, was reported to increase lifespan in mice despite dramatically lower cytochrome oxidase (COX) activity. Consistent with this, our previous studies found advantageous changes in metabolism (reduced adiposity, increased insulin sensitivity, and mitochondrial biogenesis) in Surf1 −/− mice. The lack of deleterious phenotypes in Surf1 −/− mice is contrary to the hypothesis that mitochondrial dysfunction contributes to aging. We found only a modest (nonsignificant) extension of lifespan (7% median, 16% maximum) and no change in healthspan indices in Surf1 −/− vs. Surf1 +/+ mice despite substantial decreases in COX activity (22%–87% across tissues). Dietary restriction (DR) increased median lifespan in both Surf1 +/+ and Surf1 −/− mice (36% and 19%, respectively). We measured gene expression, metabolites, and targeted expression of key metabolic proteins in adipose tissue, liver, and brain in Surf1 +/+ and Surf1 −/− mice. Gene expression was differentially regulated in a tissue‐specific manner. Many proteins and metabolites are downregulated in Surf1 −/− adipose tissue and reversed by DR, while in brain, most metabolites that changed were elevated in Surf1 −/− mice. Finally, mitochondrial unfolded protein response (UPRmt)‐associated proteins were not uniformly altered by age or genotype, suggesting the UPRmt is not a key player in aging or in response to reduced COX activity. While the changes in gene expression and metabolism may represent compensatory responses to mitochondrial stress, the important outcome of this study is that lifespan and healthspan are not compromised in Surf1 −/− mice, suggesting that not all mitochondrial deficiencies are a critical determinant of lifespan.
1. INTRODUCTION
It has been proposed that compromised mitochondrial function is a causal determinant of aging and age‐related disease. While this is a reasonable assumption, the data to support a universal direct link between reduced mitochondrial function and lifespan surprisingly do not exist. For example, we have previously shown that heterozygous (Sod2 +/−) mice with reduced levels of MnSOD, a mitochondrial antioxidant enzyme, have compromised mitochondrial function but no reduction in lifespan (Mansouri et al., 2006; Van Remmen et al., 2003; Williams et al., 1998). In contrast, a study by Dell'agnello et al. (2007) reported that mice lacking expression of the Surf1 gene, which codes for the SURF1 assembly protein for complex IV (cytochrome c oxidase, COX) of the electron transport chain (Zhu et al., 1998), show a significant increase in mean lifespan despite a dramatic reduction in COX activity (Dell'agnello et al., 2007). These studies support the concept that mitochondrial function and lifespan are not causally linked.
In humans, the SURF1 mutation leads to COX deficiency (>90%) in multiple tissues and causes a fatal neurological disorder, Leigh syndrome (Tiranti et al., 1998). Mice lacking active SURF1 protein (Surf1 −/− mice) were developed to study Leigh Syndrome (Dell'agnello et al., 2007). However, despite reduced COX activity in all tissues, the Surf1 −/− mice exhibited an unexpected increase in lifespan and a number of phenotypes that do not seem consistent with reduced mitochondrial ETC activity. For example, Surf1 −/− mice are protected from kainic acid‐induced neuronal damage, and isolated neurons from Surf1 −/− mice are resistant to cytotoxic effects of glutamate (Dell'agnello et al., 2007). Studies from our laboratory have shown that Surf1 −/− mice have reduced fat mass, improved insulin sensitivity, and enhanced memory, compared to wild‐type (Surf1 +/+) mice (Deepa et al., 2013; Lin et al., 2013). In addition, tissues from Surf1 −/− mice show increased mitochondrial biogenesis and induction of the mitochondrial unfolded protein response (UPRmt), an evolutionarily conserved stress response pathway (Baker, Tatsuta & Langer, 2011). Fibroblasts from Surf1 −/− mice also show increased resistance to cellular stresses (Pharaoh et al., 2016; Pulliam et al., 2014) suggesting that reduced COX activity induces compensatory mitochondrial stress response pathways.
To better understand how the loss of a key mitochondrial protein and dramatically reduced ETC Complex IV activity can generate the positive phenotypes listed above, we set out to critically assess the effect of Surf1 deletion on lifespan in mice and to determine the physiological changes in response to reduced mitochondrial ETC that may contribute to increased lifespan. We repeated the lifespan analysis because (i) the original lifespan study was not intended to be an aging study and did not measure maximum lifespan in all cohorts in the study; (ii) the Surf1 +/+ mice had a short lifespan relative to previously reported values for this strain (B6D2F1); and (iii) because dietary restriction (DR) has been shown to increase lifespan, we were interested in testing whether DR would further increase lifespan in the Surf1 −/− mice. Our data confirm that the Surf1 −/− mice are able to avoid a reduction in lifespan despite significant loss of COX activity. These data provide evidence that reduced mitochondrial ETC activity is not a de facto negative determinant of lifespan, and one cannot assume that all deficiencies in mitochondrial proteins will translate to negative effects on lifespan.
2. RESULTS
2.1. COX activity is reduced in Surf1 −/− mice
Surf1 deficiency was previously reported to lead to a reduction in COX activity, without affecting activity of other ETC complexes (Dell'agnello et al., 2007; Pulliam et al., 2014). Here, we measured COX activity in liver, heart, and white adipose tissue (WAT) of young (8–12 months) and old (24–27 months) Surf1 −/− and Surf1 +/+ female mice using tissue homogenates. COX activity is reduced in all three tissues ranging from 22% to 87% in young and old Surf1 −/− mice, compared to Surf1 +/+ mice on either an AL or DR diet (Figure 1 and Table 1).
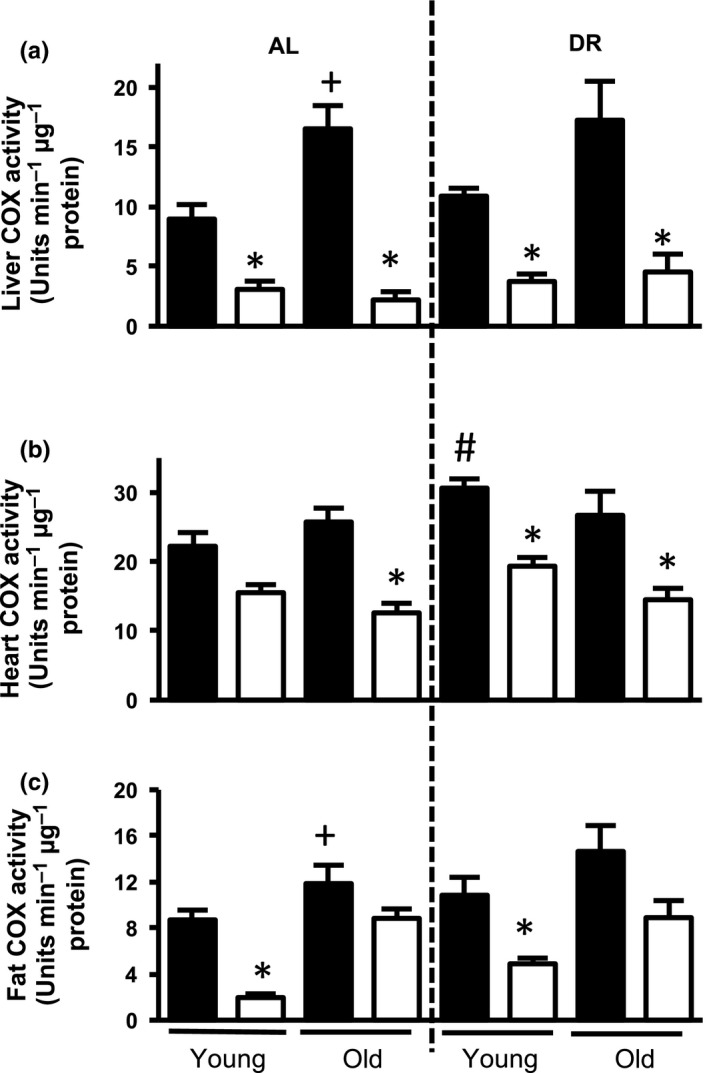
COX activity is reduced in Surf1 −/− mice fed AL or a DR diet. COX activity in tissue homogenates of liver (a), heart (b), and WAT (c) of Surf1 +/+ (black bars) and Surf1 −/− mice (white bars) fed AL or DR diet. Error bars represent mean ± SEM (n = 4–7). * Surf1 +/+ ‐AL vs. Surf1 −/− ‐AL, + young vs. old; #AL vs. DR. */+/# p < .05
Table 1
COX activity is reduced in Surf1 −/− mice fed AL or DR diet. COX activity from Figure 1 represented as percentage decrease in Surf1 −/− mice compared to Surf1 +/+ mice
| % Decrease in Surf1 −/− mice compared to Surf1 +/+ mice | ||||
|---|---|---|---|---|
| AL | DR | |||
| Young | Old | Young | Old | |
| Liver | 65.6 ± 14.6 | 86.7 ± 27.1 | 65.6 ± 10.9 | 73.7 ± 24.2 |
| Heart | 30.3 ± 2.2 | 51.0 ± 5.6 | 36.8 ± 2.4 | 45.7 ± 5.2 |
| Fat | 77.4 ± 12.6 | 25.4 ± 2.4 | 54.9 ± 5.6 | 39.2 ± 6.5 |
2.2. Surf1 −/− mice lifespan and healthspan
We measured lifespan in Surf1 +/+ and Surf1 −/− female mice fed an AL or 40% DR diet (Figure 2a, Table 2). Surf1 −/− mice fed AL showed a marginal increase in median lifespan (6.6%, p = .05) as well as nonsignificant increases in mean (3.5%, p = .46) and maximal lifespan (15.9% p = .72) compared to Surf1 +/+ mice fed AL. DR robustly extended lifespan of both Surf1 +/+ and Surf1 −/− mice, although to a lesser extent in Surf1 −/− mice. Analysis of survival curves using the Kaplan–Meier log‐rank test showed no significant difference in the survival of Surf1 −/− mice vs. Surf1 +/+ mice fed AL (p = .44) or a DR diet (p = .27), and confirmed the significant effect of the DR diet on survival. Gompertz analysis of survival is shown in Figure 2b. The Gompertz plot shows the log hazard function for each curve, modeled as a linear function of age. Both genotypes fed a DR diet showed a lower line slope, suggesting a slower age acceleration in response to DR. However, no difference was observed between Surf1 +/+ and Surf1 −/− mice on either diet. Although both Kaplan–Meier log‐rank test and Gompertz analysis failed to show a statistically significant change in lifespan between Surf1 +/+ and Surf1 −/− mice, we did detect small differences when we analyzed the difference in quartile survival time, between pairs of curves, for all quartiles (Q). Interestingly, Surf1 −/− mice fed AL showed a significant increase in lifespan at Q.5 and Q.7, compared to Surf1 +/+ mice fed AL and Surf1 −/− mice fed a DR diet showed a significant decrease in lifespan at Q.4 (p = 1.5 × 10−2) compared to Surf1 +/+ mice fed a DR diet (Table S1). Our findings show that deficiency of Surf1 does not affect overall lifespan.
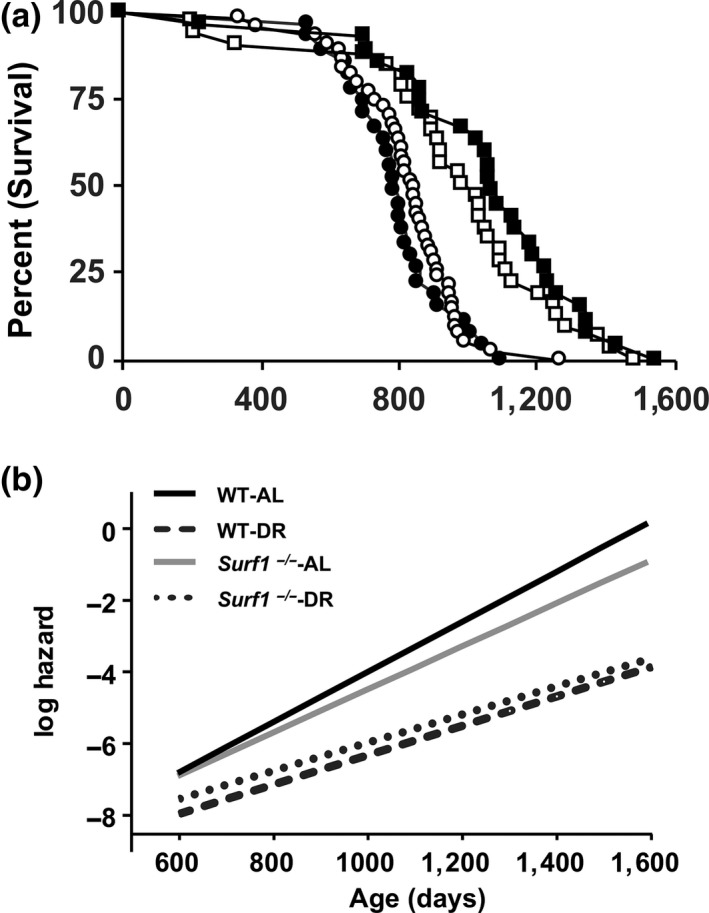
Surf1 −/− mice have increased median lifespan when fed AL, and DR reduced median lifespan of Surf1 −/− mice. (a) The Kaplan–Meier survival curves for Surf1 +/+and Surf1 −/− female mice fed AL and DR diet. The curves represent Surf1 +/+ mice fed AL (black circle, n = 29), Surf1 −/− mice fed AL (white circle, n = 32), Surf1 +/+ mice fed a DR diet (black square, n = 45), Surf1 −/− mice fed a DR diet (white square, n = 44). (b) Gompertz plot for Surf1 +/+and Surf1 −/− female mice fed AL and DR diet
Table 2
Lifespan analysis of Surf1 +/+ and Surf1 −/− mice fed AL or DR diet. The survival data from Figure 2 are expressed in days. Mean, median, 90% and maximum lifespan of Surf1 −/− mice are compared to Surf1 +/+ mice in AL and DR groups
| Surf1 +/+ ‐AL | Surf1 −/− ‐AL | Surf1 +/+ ‐DR | Surf1 −/− ‐DR | |
|---|---|---|---|---|
| Mean ± SEM | 797 ± 27.7 | 825 ± 21.5 | 1063 ± 53.8 | 973 ± 53.6 |
| Median | 794 | 850a | 1,077 | 1,009 |
| 90% | 999 | 972 | 1,352 | 1,288 |
| Maximum | 1,098 | 1,273 | 1,600 | 1,486 |
Lifespan is a valuable measure of the ultimate cumulative effects of aging (Fischer et al., 2015). However, it has become increasingly evident that changes in markers that indicate the overall general health and vitality of an organism, that is healthspan, are also highly informative (Richardson et al., 2016). To assess the effects of complex IV deficiency in the Surf1 −/− mice on healthspan, we measured several physiologic parameters including grip strength, rotarod performance, sleep/wake patterns, spontaneous cage activity in 28‐month‐old female mice and cardiovascular function in 20‐month‐old female mice. We found no significant changes in any of these parameters between Surf1 +/+ and Surf1 −/− mice (Figure 3).
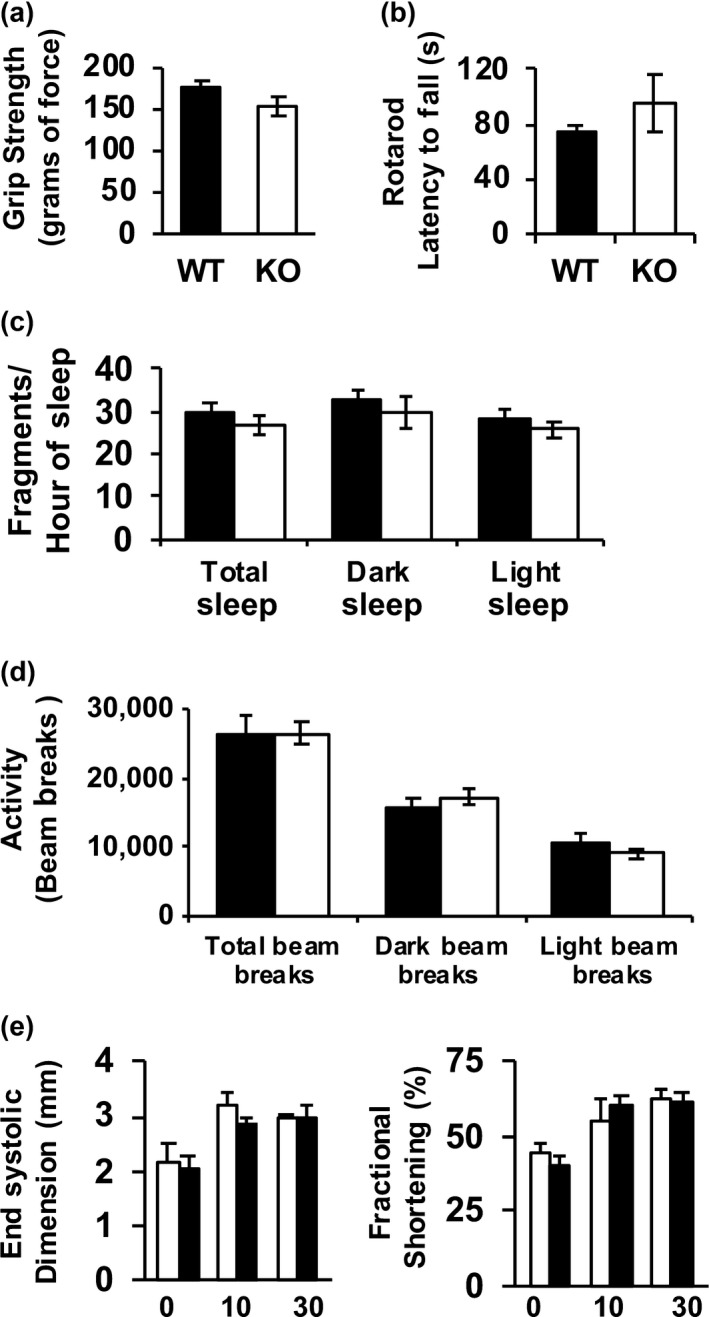
Surf1 −/− mice do not show changes in markers of healthspan. (a) Grip strength measured in 28‐month‐old Surf1 +/+and Surf1 −/− female fed AL. The grip force (in grams) was measured over 10 trials and the maximum force was recorded. (b) Rotarod performance measurement in 28‐month‐old Surf1 +/+ (black bars) and Surf1 −/− (white bars) female fed AL. The average latency to fall from the rod (in seconds) was recorded. (c) Sleep fragmentation in 28‐month‐old Surf1 +/+ (black bars) and Surf1 −/− (white bars) female fed AL expressed as the number of sleep bouts per hour of sleep (any period of inactivity (no beam breaks) greater than or equal to 40 s). (d) Spontaneous activity measured in AL‐fed 28‐month‐old Surf1 +/+ (black bars) and Surf1 −/− (white bars) mice measured as the number of beam breaks (e) Cardiovascular functions end systolic dimension (left panel) and fractional shortening (right panel) in 20‐month‐old Surf1 +/+ (black bars) and Surf1 −/− (white bars) female mice measured as previously described (Pulliam et al., 2014). Error bars represent mean ± SEM
2.3. Surf1 −/− mice fed AL but not a DR diet have a reduction in body weight and fat mass with age
Previously, we reported reduced fat mass and body weight in young Surf1 −/− male mice compared to Surf1 +/+ mice (Deepa et al., 2013). We found a similar pattern in loss of fat mass in this study. At 800 days of age, Surf1 −/− mice have 25% reduction in body weight compared to age‐matched Surf1 +/+ mice (Figure 4a). In mice fed DR, Surf1 +/+ and Surf1 −/− mice lost fat mass in a similar pattern (Figure 4b). Both genotypes on a DR diet showed similar changes in lean mass with age, however, at 800 days of age, Surf1 −/− mice fed AL have a 15% lower lean mass than age‐matched Surf1 +/+ mice fed AL (Figure 4c).
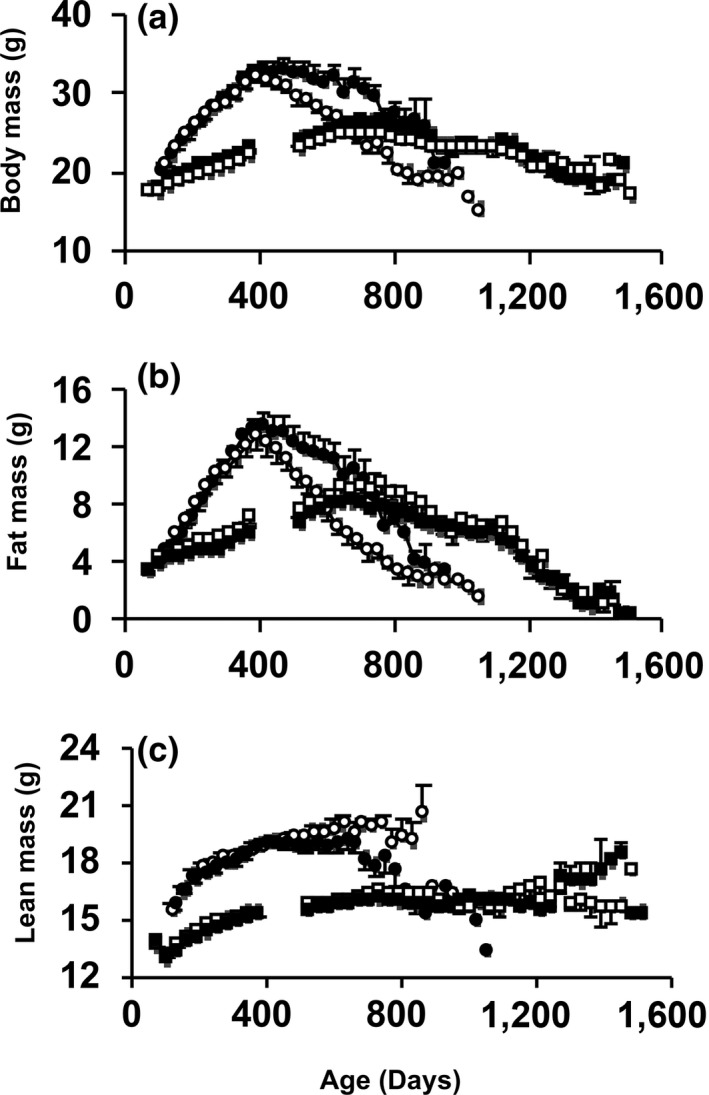
Surf1 −/− mice fed AL, not a DR diet, have a reduction in body weight and fat mass with age. Change in body weight (a), fat mass (b), and lean mass (c) with age in Surf1 +/+ and Surf1 −/− mice in the survival cohorts. The curves represent Surf1 +/+ mice fed AL (black circle), Surf1 −/− mice fed AL (white circle), Surf1 +/+ mice fed a DR diet (black square), Surf1 −/− mice fed a DR diet (white square). Error bars represent mean ± SEM
2.4. Tissue‐specific differential regulation of pathways and genes in Surf1 −/− mice
Microarray analysis was performed as an unbiased approach to compare gene expression of liver, WAT, and brain (cortex) of Surf1 +/+ and Surf1 −/− mice fed AL. We used a p < .05 cutoff to detect expression of genes significantly altered by Surf1 deficiency. In liver from Surf1 −/− mice, 714 transcripts showed a significant change compared to Surf1 +/+ mice: 336 genes were downregulated and 378 genes were upregulated (Figure 5a). In WAT of Surf1 −/− mice, 2,311 transcripts showed significant changes compared to Surf1 +/+ mice: 1,179 genes were downregulated and 1,132 genes were upregulated. In brain from Surf1 −/− mice, 318 transcripts showed significant changes compared to Surf1 +/+ mice, 119 genes were downregulated and 199 genes were upregulated. However, none of the downregulated genes were the same among the three tissues studied. In contrast, among the upregulated transcripts, ADAM Metallopeptidase Domain 11 (Adam11) and Transmembrane and Coiled‐Coil Domains 3 (Tmco3) were upregulated in all three tissues. Transcripts in liver and WAT of Surf1 −/− mice showed an overlap: 36 of the downregulated genes and 45 of the upregulated genes were similar in liver and WAT. Similarly, 11 of the downregulated genes and 23 of the upregulated genes in Surf1 −/− mice were similar in WAT and brain. Comparison of transcripts in brain and liver showed that nine of the genes are downregulated and two of the genes upregulated in both brain and liver (Figure 5a).
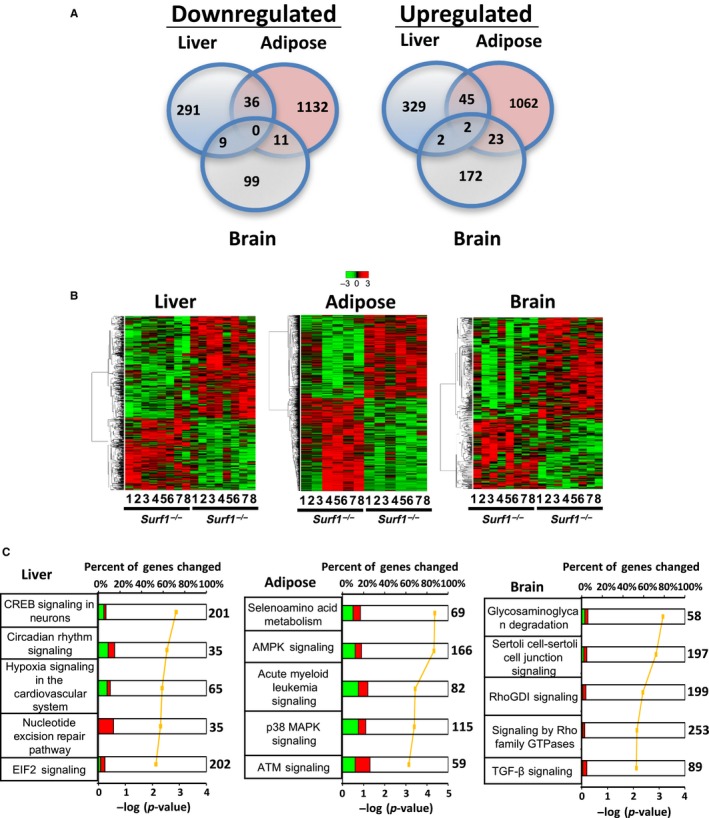
Tissue‐specific differential expression of genes and metabolic pathways in Surf1 +/+ and Surf1 −/− mice. (a) Venn diagram showing number of genes significantly downregulated (left panel) or upregulated (right panel) in liver (blue), WAT (yellow), and brain (green) using a filtering criteria of p < .05. (b) Heatmap showing expression of all the significantly changed probes in liver (left panel), WAT (middle panel), and brain (right panel). The Surf1 +/+ and Surf1 −/− samples were clustered using average linkage hierarchical cluster with Euclidean distance using a criteria of p < .05. Green represents downregulated genes and red represents upregulated genes. (c) The top five pathways from Ingenuity Pathway Analysis ranked by the lowest p‐values as determined by the Fisher's exact test are shown for liver (left panel), WAT (middle panel), and brain (right panel). The yellow line represents the ‐log of the p‐value and the bolded number on the right side of the graph represents the total number of possible genes of that pathway. Green color indicates percentage of downregulated genes, red color indicates percentage of upregulated genes, and white color indicates the percentage not found in the significant gene list. Table S2 lists the genes identified by Ingenuity Pathway Analysis
Heatmap analysis of genes that showed a significant change compared to Surf1 +/+ mice is shown in Figure 5b. Using ingenuity pathway analysis (IPA), we found that in liver, WAT, and brain 21, 95, and 30 pathways, respectively, are altered due to Surf1 deficiency. Figure 5c shows the top 5 canonical pathways that are changed in liver, WAT, and brain of Surf1 −/− mice. Even though we found significant changes in pathways in liver, WAT, and brain of Surf1 −/− mice, there was no overlap between the pathways in the three tissues. This suggests that Surf1 deficiency has a differential effect on metabolism/signaling pathways in different tissues. Table S2 shows the genes upregulated or downregulated in liver, WAT, and brain of Surf1 −/− mice.
2.5. Effect of diet and genotype on tissue metabolites in Surf1 −/− mice
A total of 221 metabolites were analyzed in adipose tissue of Surf1 +/+ and Surf1 −/− mice and of these 57 metabolites were increased and 46 metabolites were decreased (Table S3). Surf1 deficiency altered the level of 28 metabolites in adipose tissue in mice fed an AL diet: two were increased and the other 26 were decreased. Metabolites that change in response to Surf1 deficiency belong to the following metabolic pathways: amino acid (11 decreased); carbohydrate (two increased); lipid (nine decreased and one increased); and nucleotide (four decreased and one increased) (Table S3). In adipose tissue from Surf1 +/+ mice, 22 metabolites changed (eight increased and 14 decreased) and in Surf1 −/− mice 42 metabolites showed a significant change (39 increased and three decreased). Thus, Surf1 −/− mice showed a greater effect on metabolites in response to DR and the greatest effect was observed in metabolites in the lipid metabolism pathway (Table S3).
The liver tissue dataset is comprised of a total of 327 biochemicals of which 47 showed a significant change (39 increased and eight decreased) (Table S4). Several amino acids, including phenylalanine, glycine, tryptophan, leucine, and valine, were present at higher levels in Surf1 −/− mice livers, suggesting higher levels of protein turnover in Surf1 −/− mice livers. A striking accumulation of medium‐chain free fatty acids—12:0, 10:0, 9:0, and 8:0—as well as the acylglycine conjugate valerylglycine are symptomatic of a block in the function of the medium‐chain acyl‐CoA dehydrogenase (MCAD).
2.6. Metabolic pathway analysis in adipose tissue
Previously, we found that deficiency of Surf1 in male mice shift metabolism to increased fatty acid oxidation in peripheral tissues (Deepa et al., 2013). Here, we used a targeted quantitative proteomic approach to assess the changes in metabolic pathways in WAT. We analyzed WAT from Surf1 −/− mice and Surf1 +/+ mice fed AL and DR diet less than 400 days of age to see changes prior to fat loss. Of the 16 proteins analyzed in carbohydrate metabolism, only four proteins showed a significant difference due to Surf1 deficiency, three were downregulated (Hk1, Pygb, Taldo) and one was upregulated (Ldhb). DR did not alter the expression of any of the proteins in the carbohydrate metabolism panel in Surf1 +/+ mice compared to AL‐fed Surf1 +/+ mice, however, 14 proteins in Surf1 −/− mice are upregulated due to DR, compared to AL‐fed Surf1 −/− mice (Figure 6). DR also showed a genotype‐specific effect on protein expression: expression of Akr1b1, Ldhb, Slc2a4, and Taldo is increased in Surf1 −/− mice fed DR diet, compared to Surf1 +/+ mice fed DR diet.
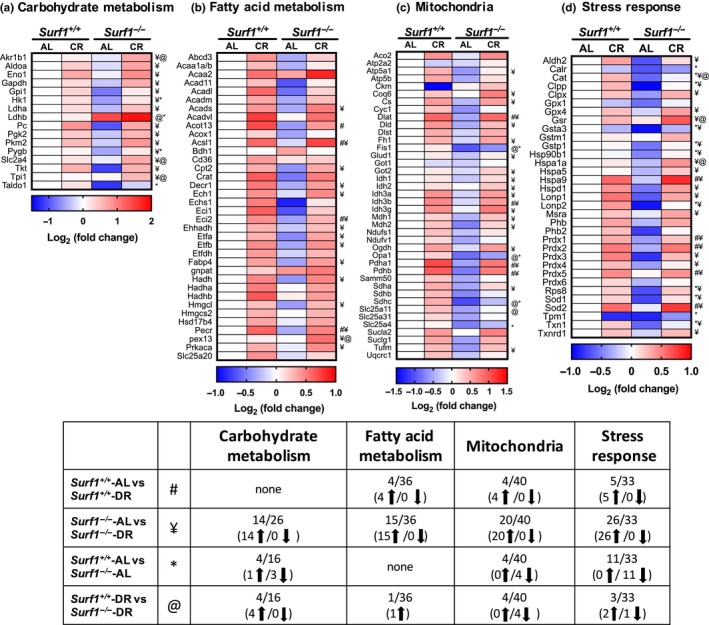
Metabolic pathways are altered in Surf1 −/− mice adipose tissue and DR reverses most of these changes. Heatmaps representing changes in protein expression of enzymes/proteins in Surf1 −/− mice compared to Surf1 +/+ mice fed AL or DR in carbohydrate metabolism (a), fatty acid metabolism (b), TCA cycle, ETC proteins, and assorted mitochondrial proteins (c), and stress response (d) in WAT obtained by targeted mass spectrometry analysis. Blue indicates downregulated proteins, white is unchanged, and red is upregulated. A summary of the data (e) represents number of enzymes/proteins in the pathway that are altered. Statistical significance determined by two‐Way ANOVA with Tukey's post hoc test (n = 7–8)
Protein expression analysis of a panel of 36 enzymes involved in mitochondrial and peroxisomal fatty acid oxidation pathways revealed no changes in Surf1 −/− mice fed an AL diet (Figure 6). Four of the proteins in the fatty acid oxidation panel were upregulated in Surf1 +/+ mice and 15 are upregulated in Surf1 −/− mice on DR. Pex13 is the only protein that showed a significant increase in DR‐fed Surf1 −/− mice compared to DR‐fed Surf1 +/+ mice (Figure 6).
Analysis of a panel of 40 enzymes/proteins in tricarboxylic acid cycle (TCA), electron transport chain (ETC), and mitochondrial enzymes/proteins showed that protein expression of four proteins was downregulated by Surf1 deficiency (Fis1, Opa1, Sdhc, and Slc25a4) (Figure 6). Twenty of the 40 proteins analyzed in the mitochondrial panel were upregulated in Surf1 −/− mice on DR diet, whereas only four proteins were upregulated in Surf1 +/+ mice fed the DR diet (Figure 6). Surf1 −/− mice fed DR diet showed downregulation of four proteins (Fis1, Opa1, Sdhc, and Slc25a11) compared to Surf1 +/+ mice fed DR diet. Therefore, Fis1, Opa1, and Sdhc were downregulated in Surf1 −/− independent of diet.
Among antioxidants, chaperones, heat shock proteins, and proteases (a total of 33 proteins) in the stress response panel, absence of Surf1 reduced the expression of 11 proteins in Surf1 −/− mice compared to Surf1 +/+ mice (Figure 6). DR increased the expression of 25 proteins in Surf1 −/− mice fed DR vs. AL diet, whereas only five proteins were elevated in Surf1 +/+ mice fed DR diet. DR also showed a genotype‐specific effect on protein expression in the mitochondrial panel: Gsr and Hspa1a were increased and catalase was decreased in Surf1 −/− mice fed DR diet, compared to Surf1 +/+ mice fed DR diet (Figure 6, Table S5).
2.7. UPRmt‐associated proteins and mitochondrial ETC complex subunits are induced transcriptionally in Surf1 −/− mice liver, but not in heart
Because the effect of aging on the UPRmt has not been reported in mice, we measured transcript and protein levels of UPRmt‐associated proteins ClpP, LonP1, and Hsp60 in young and old Surf1 −/− and Surf1 +/+ mice. In young Surf1 −/− mice liver, transcript levels of ClpP, LonP1 are elevated by 1.4‐ and 2.9‐fold, respectively, compared to young Surf1 +/+ mice (Figure S1a). However, an induction in mRNA levels for UPRmt‐associated proteins is not observed in Surf1 −/− mice heart (Figure S1b), suggesting a tissue‐specific effect of UPRmt in response to Surf1 deficiency. Our findings suggest that UPRmt‐associated proteins are induced only when the mice are young, not old, and they also show a tissue‐specific difference in their expression with age. The absence of UPRmt induction, assessed by the transcript levels of UPRmt‐associated proteins, in old Surf1 −/− mice suggests that the UPRmt is not directly involved in lifespan extension. However, the effect of UPRmt induction at young age on lifespan is not known.
Surf1 −/− male mice are reported to have elevated mitochondrial biogenesis and increased expression of ETC subunits (Pulliam et al., 2014). Here, assessment of transcript levels of ETC subunits in liver of female Surf1 −/− and Surf1 +/+ mice showed that transcript levels of ETC subunits NDUFS3 (3.8‐fold), ND1 (2.2‐fold), SDHA (3.9‐fold), SDHB (3.2‐fold), Reiske protein (3.5‐fold), COX2 (2.3‐fold), and ATPase 6 (1.6‐fold) were significantly elevated in young Surf1 −/− mice (Figure S1e). However, there was no significant difference in the transcript levels of the ETC proteins in old Surf1 −/− mice liver, compared to old Surf1 +/+ mice (Figure S1e). In contrast to the changes in ETC subunits in liver, ETC subunit mRNA levels were similar in Surf1 +/+and Surf1 −/− mice heart at young and old ages (Figure S1f).
3. DISCUSSION
The key finding of this study is that a 22%–87% reduction in activity of a mitochondrial ETC complex (Complex IV) across all tissues measured does not significantly limit lifespan or negatively affect healthspan. Further, we show numerous and tissue‐specific changes in metabolism, protein, and gene expression in response to the loss of Surf1. For many years, there has been a widely held hypothesis that mitochondrial function decreases during aging and contributes to aging and age‐related physiologic decline. Specifically, the Mitochondrial Theory of Aging predicts that damage to the mitochondrial ETC and mitochondrial DNA over time accumulates and contributes to a spiral of compromised mitochondrial function that facilitates aging (Harman, 1972). The data we present here do not support this hypothesis. Rather, our results support the concept of a threshold effect for loss of mitochondrial function and provide clear evidence for a link between the ETC and metabolism.
Despite the commonly held concept that mitochondrial dysfunction is directly related to limited lifespan, the evidence available to support this is not unequivocal. For example, reduced ETC activity can lead to increased lifespan in C. elegans mitochondrial ETC (Mit) mutants (Munkácsy & Rea, 2014). Specifically, RNAi knock‐down of genes encoding subunits of ETC complexes I, III, or IV, and mitochondrial enzyme CoQ7 (Clk‐1) results in lifespan extension (Dillin et al., 2002; Lee et al., 2003; Munkácsy & Rea, 2014). Similar to the findings in C. elegans, inhibition of all ETC complexes, except complex II, extended lifespan in flies (Copeland et al., 2009). These studies in invertebrates are in clear contrast to the idea that mitochondrial function limits lifespan and the mechanisms responsible have not been defined. In mice, the effect of compromised mitochondrial ETC on lifespan is varied. Mice heterozygous for the Mclk gene, the homolog of the C. elegans clk1 gene (the first reported long‐lived mitochondrial mutant in C elegans), have an extended lifespan (Lapointe, Stepanyan, Bigras & Hekimi, 2009), while Mclk homozygous null mice show a dramatically shortened lifespan (Hughes & Hekimi, 2011). Likewise, mice with a homozygous deletion of an ETC complex I subunit (Ndufs4 knockout mice) also show reduced longevity. Both of these mutant mouse models are very short‐lived and have severe respiratory and metabolic phenotypes (Kruse et al., 2008; Wang, Oxer & Hekimi, 2015). One explanation for the seemingly contradictory effects of ETC mutation and longevity is a threshold effect in which compromised mitochondrial function alters cellular function only once a critical deficiency in ATP production in reached. In the context of mitochondrial mutations that cause mitochondrial disease, the variable phenotypes that occur have been tied to a critical threshold of mutation load (approximately 80%–90%) above which the phenotypes associated with disease occur (Rossignol et al., 2003). For example, heterozygous knock‐in mice with reduced levels of DNA polymerase γ, a mitochondrial DNA proofreading enzyme, have a 30‐fold increase in mitochondrial DNA mutation load but show no deleterious phenotypes and have a normal lifespan while homozygous null mutants have a dramatic reduction in lifespan (Vermulst et al., 2007). Using cybrids containing a Complex I (ND5) mutation, Bai, Shakeley and Attardi (2000) showed that reduced mitochondrial respiration occurred only after the percentage of heteroplasmy reached 60%. Likewise, other studies have shown that inhibition of ETC complexes will not affect respiration or ATP production until a threshold is exceeded. In fact, this biochemical threshold was shown to be at 75% inhibition for COX in an in vitro analysis using isolated muscle mitochondria in vitro (Letellier, Heinrich, Malgat & Mazat, 1994).
Consistent with the threshold concept, we tested mitochondrial function in the Surf1 −/− mice using standard in vitro assays and found no loss of mitochondrial function and no evidence for increased ROS generation (Pulliam et al., 2014). It is possible that this is somehow related to an increase in mitochondrial biogenesis to compensate for the reduction in COX activity in tissues of the Surf1 −/− mice and inhibit any pathophysiological consequences at the cellular level. However, there is evidence that the mitochondria in the Surf1 −/− mice are impaired to some extent in vivo. For example, the Surf1 −/− mice have a significantly diminished treadmill endurance capacity and elevated blood lactate, consistent with impaired mitochondrial function (Pulliam et al., 2014).
The goal for this study was to determine the metabolic changes that occur in vivo in response to the reduction in SURF1 and Complex IV activity that might contribute to the increase in lifespan in Surf1 −/− mice originally reported by Dell'agnello et al. (2007). We were also interested in whether DR could extend this lifespan extension. Thus, we repeated the lifespan study in female mice under AL and DR (40% restriction) conditions. However, our lifespan analysis revealed only a slight and nonsignificant increase (7%) in median lifespan, compared to the 17% increase previously reported for female Surf1 −/− mice. Despite the discrepancy in the survival results, the critical fact remains that we did not see compromised lifespan despite the severe reduction in complex IV activity. This is not the first time a study was unable to replicate the increase in lifespan shown in an original study. For example, knockout mouse models for the signaling molecule p66Shc (p66shc−/− mice) (Migliaccio et al., 1999), insulin receptor substrate 2 (Irs2) (Irs2+/− mice) (Taguchi, Wartschow & White, 2007), and the IGF1 receptor (Igf1r +/− mice) (Holzenberger et al., 2003) all showed substantial lifespan extension in initial reports that was not confirmed in follow‐up studies (Bokov et al., 2011; Ladiges et al., 2009; Liang et al., 2003 ; Ramsey et al., 2014; Selman, Lingard, Gems, Partridge & Withers, 2008; Selman et al., 2007; Unnikrishnan, Deepa, Herd & Richardson, 2017). Differences in genetic background can potentially modulate lifespan results. However, this was not a contributing factor in our results as the mice in our study and the study by Dell'agnello were on the same genetic background (B6D2F1/J). In contrast, a key difference between the lifespan results in the Surf1 −/− mice lifespan study by Dell'agnello et al. (2007) and our study is the mean lifespan of the Surf1 +/+ mice. They reported a mean lifespan of 700 days for female Surf1 +/+ mice. This is considerably lower than previously reported for female Surf1 +/+ mice on B6D2F1/J background, that is, a mean lifespan 826 days (Yamate, Tajima, Kudow & Sannai, 1990). Hence, it is possible that the reduced lifespan of Surf1 +/+ mice contributed to the observed lifespan extension in Surf1 −/− mice. The Surf1 +/+ mice in our cohort show a mean lifespan consistent with previously reported values (797 days), while the Surf1 −/− mice in our cohort had a mean survival of 825 days. Interestingly, this is very close to the lifespan originally reported by Dell'agnello et al. (2007) (820 days), further supporting the possibility that the reduced Surf1 +/+ lifespan contributed to the lifespan extension they reported. We found that DR extended lifespan in both Surf1 +/+ and Surf1 −/− mice; however, the effect was surprisingly less in the Surf1 −/− mice (18% and 17% extension in mean and maximum lifespan, respectively, in the Surf1 −/− mice vs. a 33% and 46% extension in Surf1 +/+ mice). Our metabolic and phenotypic data unfortunately do not reveal a plausible explanation for the reduced response of the Surf1 −/− mice to DR, but one hypothesis might be that mitochondrial function may be more adversely affected by lower nutrient flux in response to DR in the mice with compromised COX activity.
Our previous studies in young adult male Surf1 −/− mice showed interesting metabolic phenotypes such as reduced fat mass, increased insulin sensitivity, and increased mitochondrial biogenesis in some tissues. There was also an induction of the mitochondrial unfolded protein response (UPRmt) in several tissues, suggesting that the misassembly of Complex IV subunits due to the lack of the SURF1 assembly factor might induce upregulation of proteostasis and other protective mechanisms that may be related to the increase in lifespan. In the current study, we did not find a clear induction of the UPRmt. One possible reason for this is that our original studies were performed using male mice and the current study is in female mice. We also did not find a clear relationship between the UPRmt and aging in this study. In addition, the most consistent finding in all the metabolic data we collected is the fact that COX deficiency affects gene expression metabolites and metabolic pathways in a tissue‐specific manner. Likewise, DR shows tissue‐specific effects on proteins and metabolites in Surf1 +/+ and Surf1 −/− mice. In the current study, our metabolic studies mainly focused on adipose tissue, liver and, brain. Adipose tissue and liver play a role in metabolism, and the Surf1 −/− mice tend to have less fat and improved insulin sensitivity (Deepa et al., 2013). We measured changes in brain, because many genetic manipulations restricted to brain have been shown to affect lifespan. For example, there is a report showing that a CNS‐restricted knockout of Surf1 can extend lifespan in fruit flies (Zordan et al., 2006). Adipose tissue had by far the most changes in gene expression. Over 2,000 genes were differentially regulated in adipose tissue between Surf1 +/+ and Surf1 −/− mice. These changes may be related to the dramatic loss of fat mass that occurs in the Surf1 −/− mice over their lifespan. Metabolomic analysis in adipose tissue showed 28 of 221 metabolites were different between the Surf1 +/+ and knockout mice on AL diet but only half as many (11 metabolites) differed in Surf1 +/+ vs. Surf1 −/− mice on DR. This may be related to the fact that there is no differential loss of fat mass in the Surf1 −/− mice on DR and may also be related to the reduced lifespan extension in the Surf1 −/− mice on DR compared to Surf1 +/+ mice on DR. In summary, the most important finding in this study is that reduced complex IV activity throughout the lifespan does not negatively impact lifespan or healthspan. The long held idea that mitochondrial dysfunction is a negative determinant of aging should be tempered by the idea that not all mitochondrial impairments do not accelerate aging or physiologic decline and may in fact have hidden beneficial effects on metabolism.
4. EXPERIMENTAL PROCEDURES
4.1. Animals and lifespan
Surf1 −/− mice were generated as described by Dell'agnello et al. (2007). Experiments were conducted in female Surf1 −/− B6D2F1/J (C57BL/6J_DBA/2) mice and control littermates (Surf1 +/+), unless otherwise stated, and were performed according to the protocol approved by the IACUC at the University of Texas Health Science at San Antonio. Mice for the lifespan study were generated by crossing Surf1 +/− mice to each other to obtain Surf1 −/− and Surf1 +/+ mice and only females were used for the study. For the lifespan study using AL‐fed mice, we used 32 Surf1 −/− mice and 29 Surf1 +/+, and for the dietary restricted lifespan study, 44 Surf1 −/− mice and 45 Surf1 +/+were used. Mice (5 animals/cage) were housed under barrier conditions using micro‐isolator cages and were fed AL a standard NIH‐31 chow diet (Harlan Teklad, Madison, WI, USA) on a 12‐h dark‐light cycle. Dietary restriction was initiated at 2 months of age by feeding DR mice 60% of the diet (by weight, 40% restriction) consumed by the mice fed AL. For the lifespan experiments for mice AL or a DR diet, the mice were allowed to live out their natural lifespan and lifespans for mice were determined by recording the ages of spontaneous death of the mice. The mean, median, 10% (the mean lifespan of the longest‐lived 10% of animals), and maximum (the age of death for the longest‐lived mouse in the cohort) lifespans were calculated from the survival data for each group. Body composition of the mice was determined monthly by quantitative magnetic resonance (QMR, Echo Medial Systems, Houston, TX, USA). Food consumption was measured for a one week period every two weeks for 3 months and monthly for 11 months in 19‐20 mice (four cages) per genotype. There was no difference in food consumption between the Surf1 +/+ and Surf1−/− mice. Surf1−/− mice consumed 3.1 g ± 0.1 g per day and Surf1+/+ mice consumed 3.0 ± 0.0 g per day).
4.2. COX activity assay
COX activity was measured in liver, heart, and WAT homogenates as described (Spinazzi, Casarin, Pertegato, Salviati & Angelini, 2012). The rate of oxidation of cytochrome c was calculated from the change in absorbance at 550 nm. The extinction coefficient of 21.84/mM/cm was used to determine the enzymatic activity.
4.3. Microarray analysis
Microarray processing was carried out at the National Institute on Aging, Gene Expression and Genomics Unit as previously described (Fok et al., 2014).
4.4. Metabolomics
Frozen brain tissue (n = 8) and WAT (n = 8) were analyzed for metabolite levels by Metabolon (Durham, NC, USA). At the time of analysis, samples were extracted and prepared for analysis using Metabolon's standard solvent extraction method as previously described (Ghazalpour et al., 2014). Data were normalized by Metabolon and statistical analysis was performed using ANOVA with pairwise comparisons. False discovery analysis was then applied to the dataset using r package “q value” (v 1.30.0, Dabney, A and Storey, J). Metabolon imputed the data for metabolites that had samples that were not detected using the lowest detected data value available for that metabolite. We excluded all metabolites that had imputed value >20% in the groups we analyzed for the pairwise comparisons. Statistical significance was indicated at q value < 0.05 and fold change >15%.
4.5. Targeted quantitative proteomics
Selected reaction monitoring (SRM) mass spectrometry was used to quantify protein abundance as described before (Kinter et al., 2012). Bovine serum albumin (8 pmol) was added to 60 μg (as total protein) of WAT tissue homogenate as an internal standard.
4.6. Rotarod
Rotarod performance was measured as described by previously using Rotamex 4/8 (Columbus Instruments, Columbus, OH, USA) (Muller et al., 2006).
4.7. Grip strength
Hindlimb muscle strength was determined by measuring peak force using the Digital Grip Strength meter equipped with a Hind Limb Pull Bar Assembly (Columbus Instruments, Columbus, OH, USA) essentially as described before (Zhang et al., 2014). The grip force was observed over 10 trials and the maximum force was recorded.
4.8. Activity and sleep
Spontaneous activity and sleep were measured as previously described (Fischer et al., 2016). Activity is measured in the number of beam breaks. Sleep is measured using a method validated by Pack et al. (2007). A sleep bout is any period of inactivity (no beam breaks) greater than or equal to 40 s. Sleep fragmentation is characterized by measuring the number of sleep bouts per hour of sleep. Data were collected during the light and dark phase over the testing period following acclimation (24‐hr, one light and one dark phase) (Pack et al., 2007).
4.9. Statistical analyses
In general, a two‐Way ANOVA with Tukey's post hoc test was performed using graphpad prism version 7.0b (GraphPad Software, La Jolla California). Specific statistical analyses for individual datasets are included in the Figure Legends. Kaplan–Meier estimation, Gompertz parametric modeling, log‐rank testing for differences between survival curves, and Boschloo testing for difference in extreme survival were performed with specific functions implemented in the r packages survival, eha and Exact. Quantile regression implemented in the r package quantreg was used to estimate and compare mean, median, and extreme quartile survival times for each group.
AUTHOR CONTRIBUTIONS
S.S.D. analyzed data, prepared figures, and wrote manuscript; G.P. analyzed proteomic data, helped with statistics, and edited the manuscript; M.K. performed targeted quantitative proteomic analysis; V.D. performed lifespan studies and QMR data collection; W.C.F. generated heatmap and performed pathway analysis; K.R. performed real‐time qPCR and COX enzyme assays; D.P. did genotyping of mice and western blots; S.H. assisted with various assays; K.F. and V.S. ran and analyzed healthspan measures; C.G and J.D.W. performed lifespan data analysis; C.V. provided Surf1 −/− mice and gave critical comments for the manuscript; A.R. gave critical suggestions for the manuscript; and H.V.R. designed the experiments, wrote and edited manuscript.
Supporting information
ACKNOWLEDGMENTS
We wish to thank Massimo Zeviani, MRC‐MBU, University of Cambridge, Cambridge, UK for the kind gift of Surf1 −/− mice, Kevin Becker of the IRP Core (Gene Expression and Genomics Unit at NIA) for performing microarray, and Metabolon Inc. (Durham, USA) for generating the metabolomics data. We also thank Merry Lindsey for assistance in measuring cardiac function and the San Antonio Nathan Shock Center Core for assistance with the lifespan studies. This work was supported by an Ellison Medical Foundation Senior Scholar award to H.V.R. and an NIA funded R21 (AG046803) to H.V.R. H.V.R and A.R. are supported by Senior VA Research Career Scientist awards from the Department of Veterans Affairs. C.V. is supported by a MRC core grant to MBU and D.P. was supported by the training grant (T32 AG021890).
Notes
Deepa SS, Pharaoh G, Kinter M, et al. Lifelong reduction in complex IV induces tissue‐specific metabolic effects but does not reduce lifespan or healthspan in mice. Aging Cell. 2018;17:e12769 10.1111/acel.12769 [PMC free article] [PubMed] [CrossRef] [Google Scholar]