- Journal List
- PLoS One
- PMC7304595

Brd2 haploinsufficiency extends lifespan and healthspan in C57B6/J mice
Shilpa Pathak
1 Battelle Center for Mathematical Medicine, The Abigail Wexner Research Institute, Nationwide Children’s Hospital, Columbus, Ohio, United States of America
William C. L. Stewart
1 Battelle Center for Mathematical Medicine, The Abigail Wexner Research Institute, Nationwide Children’s Hospital, Columbus, Ohio, United States of America
2 Department of Pediatrics, The Ohio State University, Columbus, Ohio, United States of America
3 Department of Statistics, The Ohio State University, Columbus, Ohio, United States of America
Christin E. Burd
4 Department of Molecular Genetics, The Ohio State University, Columbus, Ohio, United States of America
5 Department of Molecular Biology and Cancer Genetics, The Ohio State University, Columbus, Ohio, United States of America
Mark E. Hester
6 Steve and Cindy Rasmussen Institute for Genomic Medicine, Abigail Wexner Research Institute at Nationwide Children’s Hospital, Columbus, Ohio, United States of America
David A. Greenberg
1 Battelle Center for Mathematical Medicine, The Abigail Wexner Research Institute, Nationwide Children’s Hospital, Columbus, Ohio, United States of America
2 Department of Pediatrics, The Ohio State University, Columbus, Ohio, United States of America
Associated Data
- Supplementary Materials
- S1 Fig: qPCR analysis of tissue and MEFs from WT and Brd2+/- mice measuring the levels of Brd2 transcript. (DOCX)pone.0234910.s001.docx (2.5M)GUID: E03880D3-CEBF-4CE6-B56F-5943C735CEDDS2 Fig: Although it is well known that caloric restriction is associated with increased lifespan in C57B6/J mice, we do not see any evidence in support of body weight differences in our HET and WT mice. (DOCX)pone.0234910.s002.docx (5.3M)GUID: 7552F47C-3CEF-412A-ABD1-6EA06769F5F2S3 Fig: Histological differences in representative liver, spleen and testis sections from age matched WT and HET mice (n = 6) at 18 months. (A & B): Hematoxylin & Eosin staining in WT liver (A) showing inflammation (thin black arrows) and more macrovascular vacuolation (thick, black arrow) in WT than HET (B). (C & D) Hematoxylin & Eosin staining in WT spleen (C) and HET(B) showing disorganization of the splenic structure in WT. (E & F) Hematoxylin & Eosin staining in WT (A) and HET(B) testes showing increased vacuolation in WT (C) as compared to HETs (D).
(DOCX)
pone.0234910.s003.docx (7.0M)GUID: 045E3AC7-7745-4148-A189-0A973BF4F928Attachment: Submitted filename: Response to Reviewers.docxpone.0234910.s006.docx (61K)GUID: 3300FCE8-4401-4FD4-A01A-A94A194CDB28
- Data Availability Statement
All relevant data are within the manuscript and Supporting Information files.
Abstract
Aging in mammals is the gradual decline of an organism’s physical, mental, and physiological capacity. Aging leads to increased risk for disease and eventually to death. Here, we show that Brd2 haploinsufficiency (Brd2+/-) extends lifespan and increases healthspan in C57B6/J mice. In Brd2+/- mice, longevity is increased by 23% (p<0.0001), and, relative to wildtype animals (Brd2+/+), cancer incidence is reduced by 43% (p<0.001). In addition, relative to age-matched wildtype mice, Brd2 heterozygotes show healthier aging including: improved grooming, extended period of fertility, and lack of age-related decline in kidney function and morphology. Our data support a role for haploinsufficiency of Brd2 in promoting healthy aging. We hypothesize that Brd2 affects aging by protecting against the accumulation of molecular and cellular damage. Given the recent advances in the development of BET inhibitors, our research provides impetus to test drugs that target BRD2 as a way to understand and treat/prevent age-related diseases.
Introduction
Inherent in the aging process is a gradual decline in physical, cognitive, and physiological capacity, an increasing risk of disease, and ultimately death. Although it is thought that aging results from the cumulative effects of molecular and cellular damage, we serendipitously discovered that a Brd2-haploinsufficient (Brd2+/-; denoted HET) mouse model we developed to study epilepsy [1–3] had a much longer lifespan compared to wild type (Brd2+/+; denoted WT) mice. In pursuing the mechanism by which BRD2, a bromodomain (BET) protein, predisposed to epilepsy [4, 5], we found that HETs, which are overtly normal [1, 3], not only have significantly longer lifespans but also show healthier-aging phenotypes, including reduced cancer incidence and improved kidney function, as compared to wildtype mice.
To date, there are only a handful of candidate longevity genes for C57B6/J mice. Of particular interest is Cisd2, which is perhaps the most well-known longevity gene in C57B6/J mice [6, 7]. Cisd2, which is chiefly concerned with the maintenance of mitochondrial integrity, increases lifespan in C57B6/J mice when overexpressed. The Cisd2-/- mouse (which is viable) has a shortened lifespan [7].
BRD2 and longevity
There are several genes and molecular processes that are known to influence longevity in mice. Many of those genes are in turn influenced by Brd2. For example, Brd2 haploinsufficiency downregulates IGF signaling [8], and IGF signaling is decreased in calorically restricted mice—a dietary intervention that increases lifespan [9, 10]. Similarly, Brd2 haploinsufficiency up-regulates genes in the Sirtuin pathway [11], and up-regulation of the Sirtuin pathway is associated with increased lifespan [12, 13]. Specifically, Sirtuin 1 (SIRT1) and its homologs regulate longevity-related processes such as DNA repair, genome stability, inflammation, apoptosis, cell cycle progression and mitochondrial respiration [14–16]. Reduced expression of Brd2 also increases p53, Nqo1, and Hmox1 expression [11], all of which reduce oxidative stress. In addition, upregulation of p53 increases genomic stability, promotes DNA repair, and increases lifespan [17, 18]. Because Brd2 haploinsufficiency is tied to multiple longevity-related genes and molecular processes, reduced expression of Brd2 could be a fundamental—and heritable—factor influencing lifespan.
BRD2 and cancer
Whereas decreased Brd2 expression is associated with longevity-promoting processes, increased Brd2 expression can promote cancer in murine hematopoietic cells and in B-lymphocytes [19]. Furthermore, work from The Cancer Genome Atlas (TCGA) shows that BRD2 expression is elevated across 32 distinct tumor types and establishes BRD2 as a promising drug target for human cancers. Also, reducing the expression of BRD2 in HeLa cells leads to a 60% increase in tumor-suppressing p53 levels [20], which supports the notion that increased BRD2 promotes cancer growth. Hence, the overexpression of BRD2 is oncogenic, whereas inhibiting the activity of BRD2 limits cancer progression. Furthermore, the overexpression of BET proteins in general promotes cancer in mice [19, 21, 22], and reduced BET expression (via BET inhibitors) is currently being tested as a treatment for cancer in both pre-clinical models and clinical trials [23–25]. Because cancer is a major cause of age-related morbidity and mortality, we hypothesize that Brd2’s reduced expression could also increase healthspan by reducing cancer risk.
This report is the first demonstration that reducing Brd2 at the genetic level in a whole animal produces the same effects described above. We describe: 1) notable differences between Brd2 heterozygous and wildtype mice in aging-related phenotypes, including cancer incidence, kidney function, lifespan, and other aging-related measures, and, 2) evidence supporting the hypothesis that Brd2-haploinsufficiency increases lifespan and healthspan, in part, through the upregulation of cytoprotective genes.
Materials and methods
All animal procedures were reviewed and approved under IACUC protocol (AR12-00016) at Nationwide Children’s Hospital, Columbus, OH.
Mouse model development and husbandry
Brd2+/- mice that express half the normal amount of Brd2 [3] were generated according to previously described methods. Briefly, we performed targeted mutagenesis in SV129 embryonic stem cells (obtained from Baygenomics). Specifically, we used a Brd2 mutant embryonic stem cell line (RRE050) containing a gene trap vector pGT01 inserted into the first intron after the translational start site. This insertion abolished the epression of endogenous Brd2. We created Brd2+/- heterozygotes that carried one normal Brd2 allele (+) and one loss-of-function Brd2 allele (-). (A total loss of Brd2 is embryonic lethal [3, 26]. These animals were then backcrossed to wild-type C57BL/6J mice for at least 10 generations to ensure a uniform genetic background. The mice were housed in a pathogen-free barrier environment for the duration of the study. Mice were kept at 72°F, with a 12/12-h light/dark cycle and had free access to water and rodent standard chow, containing 18.4% protein and 9.5% lipid. Heterozygous mice showed no difference in fecundity as compared to wildtype animals. The research team evaluated mice at least once a week, monitored the survival of each mouse daily. Additional clinical signs were monitored weekly including body weight, general body condition, pattern of respiration, dehydration, posture, movement, response to stimuli, and condition of coat hair. The husbandry staff monitored all mice during routine maintenance/health checks and contacted the attending veterinarian as needed. Trained husbandry personnel detected signs of illness/moribundness in mice (as noted above). Sick animals were immediately brought to the attention of the veterinarian and the research staff, who then evaluated whether the mice should be euthanized according to institutional animal care and use committee (IACUC) procedures. All animal procedures were reviewed and approved under our IACUC protocol (AR12-00016). Dates of birth and death were recorded, irrespective of whether an animal was found dead or euthanized.
Mouse phenotyping at time of necropsy
All deceased mice were carefully screened for tumors (e.g. head and neck, liver, and kidney) and rectal prolapse at time of necropsy. The presence/absence of cataract was noted. Inflammation-related pathologies were determined at necropsy by inspection including swollen spleen, alopecia, skin lesions (e.g. dermatitis), and hardened kidney capsules. Inflammation in the kidney and other renal pathologies were later confirmed with microscopy. Specifically, formalin-fixed, paraffin embedded kidney samples from Brd2+/− and wildtype mice were stained using routine hematoxylin and eosin, Periodic acid-Schiff (PAS), or trichrome staining procedures prior to histological analyses. Histopathology of liver, spleen, and testes was analysed using hematoxylin and eosin staining.
Mouse epigenetic aging clock
Genomic DNA was isolated from liver tissue samples from WT and HET mice using Quick DNA universal kit (Zymo Research). Evaluation of the biological age of these samples was done using the Mouse DNAge Epigenetic Aging Clock service offered by Zymo Research. This is a mouse biological age predictor based on DNA methylation levels of a small set of CpG sites [27]. This method is based on Hovrath’s aging clock [28] and uses the Simplified Whole-panel Amplification Reaction Method.
Nqo1, Hmox1 and Sirt1 mRNA expression
Given that Brd2 regulates cytoprotective genes and pathways, we looked for differential gene expression in selected cytoprotective genes. Total RNA was extracted from kidney tissue using an RNeasy mini kit (Qiagen), per manufacturer’s instructions. Extracted RNA was then treated with DNAse I (Turbo DNA free, Invitrogen). Total RNA was reverse transcribed using iScript Reverse Transcription Supermix (Bio-Rad). Quantitative RT-PCR was performed using an Eppendorf thermocycler with TaqMan Gene Expression master mix (Applied Biosystems). Relative expression of the target genes was calculated with respect to a housekeeping gene (i.e. mGapdh). Assay identification numbers are listed in Table 1.
Table 1
| Gene | IDT assay ID |
|---|---|
| Nqo1 | Mm.PT.58.10871473 |
| Hmox1 | Mm.PT.58.8600055 |
| Sirt1 | Mm.PT.58.7263242 |
| Gapdh | Mm.PT.39a.1 |
p53 immunoblotting
p53 protein expression is strongly associated with tumor suppression [29], and we see reduced tumor incidence in our HET mice relative to wildtype. This observation led us to measure p53 expression in liver. Protein was extracted from the liver tissue of Brd2 wildtype and heterozygous mice using RIPA buffer (Pierce) and total protein was quantified using BCA protein assays (Pierce). The protein samples were resolved on Mini-protean TGX stain-free gels (Bio-Rad) and transferred to a PVDF membrane. Primary antibodies directed against wild type p53 (Millipore Sigma, 1:1000) and Gapdh (loading control, Calbiochem, 1: 10,000) were used to probe the PVDF membrane and were detected by appropriate alkaline phosphatase-coupled antibodies (Santa Cruz Biotechnology). The resulting immunoblots were developed on a Typhoon FLA 9500 (GE Healthcare) imager after the addition of ECF substrate (GE Healthcare).
Statistical procedures used in the analyses
To test differences in survival between WT and HET mice, we compared standard Kaplan-Meier estimates of the survivor functions using log-rank tests. To test whether the correlation between Brd2 and longevity depends on the presence or absence of tumor, we implemented a standard bootstrap procedure within groups (wildtype with tumors: WT+, wildtype without tumors: WT-, heterozygotes with tumors: HET+, heterozygotes without tumors: HET-). Specifically, for each bootstrap resample we computed the difference in median survival between WT+ and HET+, and the difference in median survival between WT- and HET-. Then, we constructed a confidence interval for the change in the difference of medians to test the null hypothesis of independence. Lastly, t-tests were used to compare differences in gene expression between WT and HET animals.
Results
Brd2-haploinsufficient mice show increased longevity
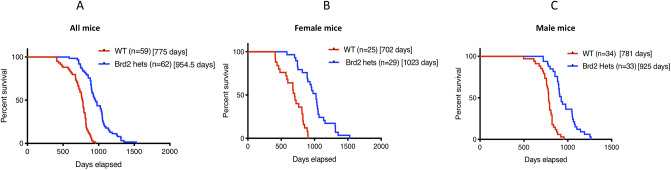
In our colony of Brd2 heterozygous and wildtype mice, Brd2+/- mice showed a significant lifespan extension as compared to WT controls (Fig 1A). The median survival of HET mice was 954.5 days, ~ 23% longer than median survival of wildtype animals (775 days; p<0.0001). The longest-lived mouse was a Brd2 heterozygous animal that survived 1,528 days, and notably, this mouse had no tumors at necropsy. Moreover, the extended lifespan of HET mice was not affected by the presence or absence of tumors at necropsy (p = 0.6), i.e., overall, HET mice with tumors lived as long as HET mice without. However, there is suggestive evidence that sex influenced the degree to which lifespan increased in Brd2+/- mice compared to wildtype, (Fig 1B and 1C; bootstrap analysis; p = 0.068). Specifically, HET females showed a 46% extension in median life span compared to WT females (1,023 vs. 702 days). In contrast, HET males showed a 19% extension in median lifespan (925 vs 781 days).
Brd2-haploinsufficient mice are biologically younger
We used Horvath’s epigenetic clock (aka the Zymo clock) to determine the biological ages of 2 WT and 2 HET mice from biopsied liver samples; all 4 mice were age-matched (770 days). HET mice showed significantly younger liver tissue than WT (Table 1; p<0.012, SEM(WT): 4.5 and SEM(HET): 7.1). The biological age of the liver samples from the HET mice suggests that HET livers are less than half the biological age of WT livers. This is corroborated by liver histology, which reveals the loss of cytoarchitecture and inflammation in WT but maintenance of liver health histologically in HETs (S3 Fig). These data provide further confirmation that Brd2 haplo-insufficiency increases lifespan and healthspan, and that it does so at a fundamental biological level.
Brd2-haploinsufficient mice exhibit delayed aging
We confirmed haploinsufficiency of Brd2 transcripts by qPCR in a broad spectrum of tissues (S1 Fig).
Necropsy data suggested that Brd2 heterozygous animals have delayed onset of aging. Because this mouse strain typically show signs of aging around 500 days, we examined the overt health of wildtype and heterozygous mice at this timepoint. At about 500 days, wildtype animals began to show symptoms of aging: reduced and slower movement, an “unkempt” appearance, hunching posture, declining health activity over the course of weeks, exhibiting an increasing decline in body condition, and slowly becoming hunched and lethargic. In contrast, HETs at the same age were youthfully active and remained healthy and fit in appearance until within about a week before death. These observations suggest that relative to WT, long-lived HET mice manifest age-related changes at a much older age and for a shorter period of time before death.
We also observed skin lesions in WT mice such as alopecia, dermatitis, and scarring at about 500 days. Age-matched HET mice showed sleek fur and no lesions. On necropsy, the elderly WT mice also had enlarged spleens (indicating chronic infection), an increased incidence of neoplastic lesions, and rectal prolapse compared to age-matched HET mice (Table 2). Next, we totaled the presence/absence of all 5 factors noted in Table 2: Dermatitis, Nephropathy, Rectal Prolapse, Lethargy and Moribund, and Ocular Lesions, (Table 2) and used this as a surrogate measure of health. Then, we performed logistic regression to test this measure for association with genotype while accounting for differences in lifespan. Based on this analysis, it is clear that HET mice have fewer pathologies by the time of death than WT mice and the difference is highly statistically significant (p<0.00011).
Table 2
| Chronological age (Days) | Biological age (DNAge) (Days) | |
|---|---|---|
| WT Female | 770 | 595 |
| WT Male | 770 | 532 |
| Het Female | 770 | 147.7 |
| Het Male | 770 | 247.1 |
Interestingly, the weights of the HETs and WT mice were comparable throughout their lives (see S2 Fig).
Pathologies present in the wildtype mice included: tumor, inflammation (including dermatitis), ocular lesions (e.g cataract, tumors, etc.), and nephropathy, which are all typical of the C57BL/6 strain (Table 3) [30]. By contrast, overt morbidities were reduced in Brd2 heterozygous animals, with the most marked decreases being in tumor incidence and nephropathology (Table 3).
Table 3
| WT | HET | |
|---|---|---|
| n (%) | n (%) | |
| Tumors | 38/59 (64.4) | 13/62 (20.96) |
| Dermatitis | 25/59 (42.37) | 10/62 (16.12) |
| Nephropathy | 32/59 (54.23) | 9/62 (14.51) |
| Rectal prolapse | 14/59 (23.72) | 10/62 (16.12) |
| Lethargy and moribund | 28/59 (47.45) | 12/62 (19.35) |
| Ocular lesions | 18/59 (30.5) | 15/62 (24.19) |
| No Apparent Pathology | 19/59 (32.2) | 36/62 (58.06) |
Fertility differences between HET and WT
Preliminary experiments examining breeding performance of our HET males indicates that HET males remain fertile longer than WT males. In a preliminary experiment involving 8 month old males, we mated 6 WT and 6 HET males to normal cycling WT females. Of the 12 WT females mated to the 6 (8 month old) WT males only 1 female was impregnated, whereas, of the 12 WT females mated to the 6 (8 month old) HET males, all 12 females were impregnated (Fisher’s Exact Test: p = 0.015). Furthermore, we mated the same 6 WT males (above) to the 12 WT females originally impregnated by the 6 HET males (ie, proven fertile WT females); and none of the 12 WT females were impregnated by the WT males, providing added confirmation that at 8 month old WT males are no longer fertile but same-aged HET males are fertile.
Overall, the above observations on health status and fecundity demonstrate that, in addition to having a longer lifespan than WT mice, HET mice also have increased healthspan.
HET mice have a lower incidence of cancer at necropsy
We observed a significant reduction in solid-tumor incidence in HET mice (13 out of 62) as compared to WTs (38 out of 59) for any solid tumor (p = 0.0000014); this reduction was independent of sex (Table 3). The spectrum of types of cancer did not differ between HET and WT mice and included tumors associated with the liver and gastrointenstinal tract, as well as head and neck tumors. Among the mice with grossly visible neoplastic disease at necropsy, HET mice with visible solid tumors (median survival 925 days) showed longer lifespan than WT mice with visible solid tumors (median survival 781 days). The reduced tumor burden at death in the HET mice suggests that they are tumor-resistant as compared to WT mice. There was reduced cancer incidence in HET mice despite the increased time for tumor growth afforded by the 23% increase in lifespan. Moreover, the effect of Brd2 haploinsufficiency in extending lifespan in HET mice is independent of the presence or absence of tumors, since HET mice with tumors lived as long as HET mice without.
Kidney pathology and function in HETs and WTs
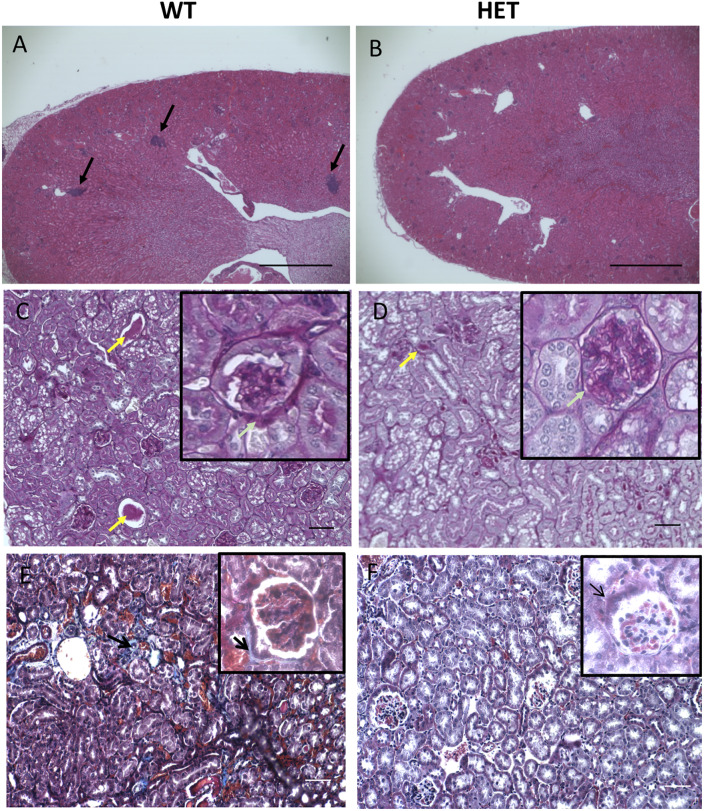
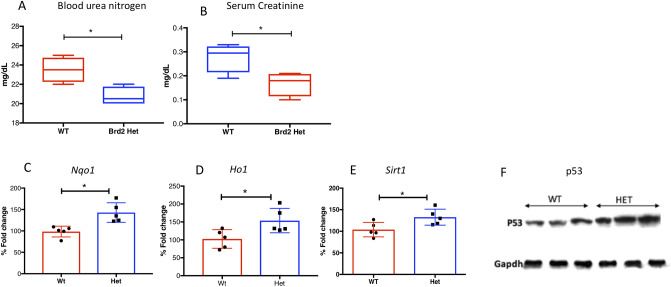
Because kidney pathology is an age-related pathology in both mice and humans, we evaluated age-related histological changes to the kidney. We observed a much lower incidence of nephropathies in HET mice than WT (Table 2). WT mice at 500 days showed the expected age-appropriate renal pathologies [31] including elevated inflammatory cell foci (often lymphocytes and neutrophils) (p = 0.0001) and tubular cytoplasmic vacuolation (Fig 2A). We observed glomerulonephropathy with hyaline material expanding the mesangium (PAS positive), dilation of renal tubules and proteinaceous casts in aged WT (Fig 2C) mice. Additionally, levels of renal fibrosis were markedly increased in aged WT mice (Fig 2E). Not only were these pathologies not seen in age-matched HET mice (Fig 2B, 2D and 2F), but HETs also maintain healthier kidney function than their age-matched WT littermates. Specifically, blood urea nitrogen and creatinine levels of HET mice were lower than that of WTs (Fig 3A and 3B).
(A & B): Hematoxylin and Eosin staining in WT (A) and HET (B) kidneys showing evidence of increased inflammatory cell foci in WT (p = 0.0001) (Black arrows) as compared to HET animals. Original magnification 4X. Scale bar; 100 μm. (C & D): Periodic acid-Schiff (PAS) staining showing larger and more numerous intraluminal proteinaceous casts in WT (C) as compared to HET (D) mice (yellow arrow). Insert shows glomerulonephropathy (thickening of Bowman’s capsule) in WT (green arrow) versus HET mice. (E & F): Masson-Trichrome staining of WT (E) and HET (F) kidney sections showing increased fibrosis indicated by the blue staining in WTs (Black arrows) as compared to HET mice. Original magnification for C, D, E and F: 20X. Scale bar; 50 μm.
Biochemical and molecular changes in aged WT and HET mice: (A-B): Blood urea nitrogen (A) and serum creatinine levels (B) in WT and HET mice (n = 6/group) showing a significant increase in WT as compared to HET animals (p < 0.05). (C-E): Steady state levels Nqo1, Hmox1 and Sirt1 mRNA (n = 5/group) in WT and HET kidneys, showing upregulation of these cytoprotective genes in samples from HET mice (p <0.05). (F): Representative immunoblot showing elevated p53 levels in HET livers (n = 6/each group).
We chose to focus on kidney-related pathologies for two reasons. First, because the protective effect of Brd2 haploinsufficiency was most pronounced in kidney (Table 3), and second, because the expression of cytoprotective genes in kidney allowed us to test our hypothesis that upregulation of cytoprotective genes leads to increased lifespan in HETs. However, in addition to the aforementioned kidney-related pathologies seen in WT mice, we also observed notable pathologies in other organs of the WT mice not seen in the HET mice. These included hyperplasia and cysts in liver, lymphoid hyperplasia in spleen, and testicular degeneration (S3 Fig).
Upregulation of cytoprotective genes in Brd2 HET kidneys
It is well-recognized that the kidney aging process is accompanied by increased oxidative stress [32]. To test the effect of cytoprotective genes in HETs versus WTs, we evaluated expression of four cytoprotective genes in kidney: heme oxygenase (HO)-1, NADPH quinone oxidoreductase 1 (Nqo1), Sirt1, and p53. We selected these genes because they are reported to be, in part, regulated by Brd2 [11, 20, 33]. The steady state mRNA levels of HO-1 and Nqo1, which are antioxidant proteins, were higher in HET kidneys as compared to WT kidneys. This suggests that the increased protection of kidney structure and function in HETs arises from the upregulation of their antioxidant defense system (Fig 3C and 3D). Sirt1, which also has a reno-protective effect [34], showed increased mRNA levels in HET kidneys as compared to WT kidneys (Fig 3E). Evaluation of p53, another cytoprotective gene, showed higher protein levels in HET kidneys compared to WT kidneys (Fig 3F). These observations support the hypothesis that Brd2 haploinsufficiency may influence longevity through the upregulation of cytoprotective genes.
Discussion
Our Brd2 haploinsufficient mouse model provides experimental evidence that reduced expression of Brd2—a gene closely tied to DNA, histones, chromatin structure, transcription, splicing, and possibly DNA methylation—can significantly alter the aging process, provide resistance to tumorigenesis, and delay the development of age-related pathologies in the kidneys. Our Brd2 haploinsufficient (HET) mice show significant lifespan extension and delays in age-related pathologies. Compared to WT mice, HETs displayed: (1) increased longevity (with or without tumors at necropsy), (2) reduced incidence of cancer at necropsy, (3) more youthful kidney structure and function, (4) increased expression of cytoprotective genes, and (5) a notable delay in age-related postural and behavioral phenotypes, (6) and extended fertility.
We hypothesize that healthspan and lifespan in humans could be increased by a reduction in BRD2 expression, in much the same way that our HETs show increased healthspan and lifespan relative to WTs. Support for this hypothesis follows from several observations. First, Brd2 is overexpressed in several human tumors (TCGA database), and overexpression of Brd2 (and other BET proteins) promotes cancer in mice [19, 21, 22]. Second, the persuit of cancer-related clinical trials for BET inhibitors [23–25, 35] suggests that Brd2 haploinsufficiency reduces cancer incidence in humans, just as our HET mice showed markedly reduced cancer incidence compared to WT (Table 3). Thus, inhibiting the activity of BRD2 in humans may increase longevity (and/or healthier aging) by reducing cancer incidence.
Interestingly, the decision to pursue Cisd2 as a candidate longevity gene was motivated by a human family study of extremely old siblings [36]. Similarly, the decision to pursue BRD2 was motivated by a family study of human epilepsy. Given that Cisd2 and Brd2 appear to have a heritable influence on longevity in humans, and given that both genes increase lifespan in C57BJ/6 mice, Cisd2 and Brd2 should be considered among the top candidate genes for longevity.
It is important to note that, while three independent mouse models show reduced levels of Brd2, the Wang et al. [37] haploinsufficient model gives rise to obese mice—a phenotype not seen in our mice [3] nor in the mice of Gyuris et al. [26]. Furthermore, the Wang et al. (2009) [37] mice also show residual Brd2 expression among BRD2(-/-) double knockout mice. By contrast, our mouse model [3]and the mouse model of Gyuris et al. (2009) [26] gave rise to the same phenotype, namely dysmorphic brain development in homozygous knockout mice. Both models used the same gene trap (ES cell RRE050); our mice also showed seizure susceptibility and changes in brain structure in HETS [1, 2].
We did not observe appreciable difference in the median lifespans of our WT males and the Jackson Lab (JAX) C57BL/6J mouse colony. In our longevity tabulations we included all mice that died before adulthood, which Jackson labs does not. Furthermore, there is notable variations in average lifespan of this strain of mice across vivaria, especially differences in deciding when and with what pathologies aging mice should be euthanized [38, 39]. Furthermore, although the salient comparison is the one between our HETs and our WTs, it is also true that our HETs live longer (on average) than Jackson Labs C57BL/6J WT mice (p<0.002) [40].
Molecular mechanisms for Brd2-related aging
As noted in Introduction, there are several speculated mechanisms for mammalian aging that have received considerable attention. However it is unlikely that any one of them (in isolation) explains Brd2’s wide-range of effects on aging. Furthermore, Brd2’s effect on aging is at a most basic level, as evidenced by the striking differences in biological ages between HET and WT age-matched mice as measured by the “methylation clock”. As such, we believe that the Brd2 HET mouse is an excellent place to start unraveling the combined effects of multiple age-related mechanisms while possibly uncovering novel mechanisms as well that could explain the differences we see in lifespan and healthspan.
The significance of cancer reduction in Brd2 HET mice
We observed a lower incidence of tumors in HET mice, suggesting that these animals are protected from developing tumors. Furthermore, we find that tumor bearing HET mice live longer, not only than tumor-bearing WT mice, but also longer than WT mice without tumors. This suggests that either the onset of tumors begins later in HET mice, or the growth of tumors is slower in tumor bearing HET mice than in tumor bearing WT mice. In our earlier work, cell cycle analysis of mouse embryonic fibroblasts derived from double knockout embryos (Brd2-/-, i.e., embryos lacking all Brd2), resulted in cell cycle delay at the G1-S transition [3]. Thus, the apparent tumor-inhibition in the HET mice could be due to Brd2’s function as an important cell cycle regulator. Because HETs live longer than WT mice whether cancer is present or not, cell cycle-related phenomena alone cannot explain the increase in longevity. Alternatively, many cytoprotective genes (e.g. p53, Sirtuin1, and NRF2) overlap with aging-related pathways that BRD2 is known to regulate [11, 20, 33]. These cytoprotective pathways include: IGF-Insulin, mTOR, p53, and DNA damage signaling. Therefore, we hypothesize that heightened expression of cytoprotective genes is crucial mechanism explaining Brd2-induced increases in healthspan and lifespan.
Oxidative stress and reduced kidney pathology
Oxidative stress is a major contributor to kidney damage [41], and increased levels of BRD2 induce oxidative stress [33] in human cells. Therefore, Brd2 haploinsufficiency, which reduces oxidative stress, could result in the healthier kidneys seen in HETs compared to WTs mice. This idea is supported by our results, and the results of others [33], showing upregulation of these antioxidants: Nqo1 and Hmox1 in HET kidneys. Our histopathological and kidney function results confirm this prediction. The youthful kidney anatomy seen in aged HET mice likely results, in part, from reduced renal oxidative stress.
Longevity genes and kidney pathology
Some reno-protective genes are also reported to longevity-related (e.g. Klotho, PPARy) [42, 43]. An example is SIRT1 [12, 44–46], whose upregulation protects the kidney by reducing inflammation [47, 48], and whose over-expression has also been associated with longevity [12]. We observed upregulation of SIRT1 and reduced inflammation in the kidneys of our long-lived HET mice. These data suggest that Brd2 haploinsufficiency promotes kidney youth by reducing inflammation through the upregulation of SIRT1 [11]. Crucially, Sirt1 is associated with anti-aging effects in hypothalamus [49], cortex, striatum [50–53], hippocampus [54, 55], as well as the kidney [56].
Chronic inflammation accelerates aging [57] and is one of the determinants of mortality [58]. It is interesting that BET proteins are critical for inflammatory response and Brd2 is essential for pro-inflammatory cytokine production [59]. Emerging preclinical and clinical evidence show a number of small molecule BET inhibitors with a potent anti-inflammatory activity [60, 61]. Thus, the reduced inflammation observed in HET kidneys could be due to Brd2’s up-regulation of cytoprotective genes when it is haplodeficient, but it could also be due to a down-regulation of pro-inflammatory genes. Extreme longevity observed in our model could be the result of either the production of anti-inflammatory cytoprotective genes or a direct effect of on inflammatory genes, but in either case, caused by Brd2 haploinsufficiency.
Taken together, our data implicate Brd2 as a gene influencing longevity and healthspan. Brd2 haploinsufficiency significantly reduces the incidence of cancer at necropsy and facilitates healthier aging. The decrease in Brd2 is positively correlated with increased cytoprotective signaling through pathways like p53-, Sirt1- and Nrf2. These pathways regulate aging-related processes such as cellular senescence, apoptosis, genome maintenance, inflammation and cellular stress mitigation. Furthermore, since Brd2 has both genetic and epigenetic functions [62], it suggests that interconnected genetic, and likely epigenetic, mechanisms are involved in the regulation of longevity. We hypothesize that the extreme longevity seen in our HET mice is mediated by Brd2 haploinsufficiency through the up-regulation of cytoprotective genes. Our results demonstrate—for the first time—a pronounced role of Brd2 in healthier murine aging, making Brd2 an excellent target for increasing both the lifespan and healthspan of mammals.
Supporting information
S1 Fig
qPCR analysis of tissue and MEFs from WT and Brd2+/- mice measuring the levels of Brd2 transcript.(DOCX)
S2 Fig
Although it is well known that caloric restriction is associated with increased lifespan in C57B6/J mice, we do not see any evidence in support of body weight differences in our HET and WT mice.(DOCX)
S3 Fig
Histological differences in representative liver, spleen and testis sections from age matched WT and HET mice (n = 6) at 18 months.(A & B): Hematoxylin & Eosin staining in WT liver (A) showing inflammation (thin black arrows) and more macrovascular vacuolation (thick, black arrow) in WT than HET (B). (C & D) Hematoxylin & Eosin staining in WT spleen (C) and HET(B) showing disorganization of the splenic structure in WT. (E & F) Hematoxylin & Eosin staining in WT (A) and HET(B) testes showing increased vacuolation in WT (C) as compared to HETs (D).
(DOCX)
Acknowledgments
The authors thank Dr. Meng Wang for the statistical analysis of data. We would like to acknowledge Ms. Emily Cameron and Mr. Vraj Patel for assistance with data collection and genotyping.
Data Availability
All relevant data are within the manuscript and Supporting Information files.
References
- PLoS One. 2020; 15(6): e0234910. »
- Decision Letter 0
Decision Letter 0
16 Jan 2020
PONE-D-19-32190
Brd2 haploinsufficiency extends lifespan and healthspan in C57B6/J mice
PLOS ONE
Dear Dr Pathak,
Thank you for submitting your manuscript to PLOS ONE. After careful consideration, we feel that it has merit but does not fully meet PLOS ONE’s publication criteria as it currently stands. Therefore, we invite you to submit a revised version of the manuscript that addresses the points raised during the review process.
As you can see from the accompanying critiques the reviewers find merit in your work but request additional information before the manuscript can be considered for publication. This relates for example to the documentation of pathology in the animal cohorts. Also, please comment on point 1 raised by reviewer 2 regarding the lifespan of the control animals. Please also address all additional points in a revised version of your manuscript.
We would appreciate receiving your revised manuscript by Feb 22 2020 11:59PM. When you are ready to submit your revision, log on to https://www.editorialmanager.com/pone/ and select the 'Submissions Needing Revision' folder to locate your manuscript file.
If you would like to make changes to your financial disclosure, please include your updated statement in your cover letter.
To enhance the reproducibility of your results, we recommend that if applicable you deposit your laboratory protocols in protocols.io, where a protocol can be assigned its own identifier (DOI) such that it can be cited independently in the future. For instructions see: http://journals.plos.org/plosone/s/submission-guidelines#loc-laboratory-protocols
Please include the following items when submitting your revised manuscript:
- A rebuttal letter that responds to each point raised by the academic editor and reviewer(s). This letter should be uploaded as separate file and labeled 'Response to Reviewers'.
- A marked-up copy of your manuscript that highlights changes made to the original version. This file should be uploaded as separate file and labeled 'Revised Manuscript with Track Changes'.
- An unmarked version of your revised paper without tracked changes. This file should be uploaded as separate file and labeled 'Manuscript'.
Please note while forming your response, if your article is accepted, you may have the opportunity to make the peer review history publicly available. The record will include editor decision letters (with reviews) and your responses to reviewer comments. If eligible, we will contact you to opt in or out.
We look forward to receiving your revised manuscript.
Kind regards,
Christoph Englert
Academic Editor
PLOS ONE
Journal Requirements:
When submitting your revision, we need you to address these additional requirements:
1. Please ensure that your manuscript meets PLOS ONE's style requirements, including those for file naming. The PLOS ONE style templates can be found at http://www.plosone.org/attachments/PLOSOne_formatting_sample_main_body.pdf and http://www.plosone.org/attachments/PLOSOne_formatting_sample_title_authors_affiliations.pdf
2. To comply with PLOS ONE submissions requirements, please provide method(s) of sacrifice in the Methods section of your manuscript.
3. As part of your revision, please complete and submit a copy of the ARRIVE Guidelines checklist, a document that aims to improve experimental reporting and reproducibility of animal studies for purposes of post-publication data analysis and reproducibility: https://www.nc3rs.org.uk/arrive-guidelines. Please include your completed checklist as a Supporting Information file. Note that if your paper is accepted for publication, this checklist will be published as part of your article.
4. Please compile all raw blot and gel images in a single PDF file titled S1_raw_images, and upload this file as Supporting Information or provide it via a publicly available data repository and include the dataset identifier (DOI or equivalent) in the Data Availability Statement. We ask that you ensure that every image in the file is clearly labeled to annotate the loading order, identity of experimental samples, method used to capture the image, and which figure panel was generated from that original image. Molecular weight markers should be included or indicated on each blot/gel image, and any lanes not included in the final figure should be marked with an “X” above the lane label on the original blot/gel image. All labeling and annotations should be performed without obscuring any data or background bands. Please note, there is a 20 MB maximum file size for Supporting Information files. If your PDF size is larger, please use a suitable repository or discuss with the journal staff.
5. Please note that all PLOS journals ask authors to adhere to our policies for sharing of data and materials: https://journals.plos.org/plosone/s/data-availability. According to PLOS ONE’s Data Availability policy, we require that the minimal dataset underlying results reported in the submission must be made immediately and freely available at the time of publication. As such, please remove any instances of 'unpublished data' or 'data not shown' in your manuscript and replace these with either the relevant data (in the form of additional figures, tables or descriptive text, as appropriate), a citation to where the data can be found, or remove altogether any statements supported by data not presented in the manuscript.
6. In your response letter, please note whether your blot/gel image data are in Supporting Information or posted at a public data repository, provide the repository URL if relevant, and provide specific details as to which raw blot/gel images, if any, are not available.
7. Thank you for your ethics statement: "All animal procedures were reviewed and approved under IACUC protocol (AR12-00016) at Nationwide Children's Hospital, Columbus, OH."
Please amend your current ethics statement to include the full name of the ethics committee that approved your specific study.
For additional information about PLOS ONE submissions requirements for ethics oversight of animal work, please refer to http://journals.plos.org/plosone/s/submission-guidelines#loc-animal-research
Once you have amended this/these statement(s) in the Methods section of the manuscript, please add the same text to the “Ethics Statement” field of the submission form (via “Edit Submission”).
8. In your Data Availability statement, you have not specified where the minimal data set underlying the results described in your manuscript can be found. PLOS defines a study's minimal data set as the underlying data used to reach the conclusions drawn in the manuscript and any additional data required to replicate the reported study findings in their entirety. All PLOS journals require that the minimal data set be made fully available. For more information about our data policy, please see http://journals.plos.org/plosone/s/data-availability.
Upon re-submitting your revised manuscript, please upload your study’s minimal underlying data set as either Supporting Information files or to a stable, public repository and include the relevant URLs, DOIs, or accession numbers within your revised cover letter. For a list of acceptable repositories, please see http://journals.plos.org/plosone/s/data-availability#loc-recommended-repositories. Any potentially identifying patient information must be fully anonymized.
Important: If there are ethical or legal restrictions to sharing your data publicly, please explain these restrictions in detail. Please see our guidelines for more information on what we consider unacceptable restrictions to publicly sharing data: http://journals.plos.org/plosone/s/data-availability#loc-unacceptable-data-access-restrictions. Note that it is not acceptable for the authors to be the sole named individuals responsible for ensuring data access.
We will update your Data Availability statement to reflect the information you provide in your cover letter.
9. PLOS ONE now requires that authors provide the original uncropped and unadjusted images underlying all blot or gel results reported in a submission’s figures or Supporting Information files. This policy and the journal’s other requirements for blot/gel reporting and figure preparation are described in detail at https://journals.plos.org/plosone/s/figures#loc-blot-and-gel-reporting-requirements and https://journals.plos.org/plosone/s/figures#loc-preparing-figures-from-image-files. When you submit your revised manuscript, please ensure that your figures adhere fully to these guidelines and provide the original underlying images for all blot or gel data reported in your submission. See the following link for instructions on providing the original image data: https://journals.plos.org/plosone/s/figures#loc-original-images-for-blots-and-gels.
In your cover letter, please note whether your blot/gel image data are in Supporting Information or posted at a public data repository, provide the repository URL if relevant, and provide specific details as to which raw blot/gel images, if any, are not available. Email us at gro.solp@enosolp if you have any questions.
10. PLOS requires an ORCID iD for the corresponding author in Editorial Manager on papers submitted after December 6th, 2016. Please ensure that you have an ORCID iD and that it is validated in Editorial Manager. To do this, go to ‘Update my Information’ (in the upper left-hand corner of the main menu), and click on the Fetch/Validate link next to the ORCID field. This will take you to the ORCID site and allow you to create a new iD or authenticate a pre-existing iD in Editorial Manager. Please see the following video for instructions on linking an ORCID iD to your Editorial Manager account: https://www.youtube.com/watch?v=_xcclfuvtxQ
11. We note that you have included the phrase “data not shown” in your manuscript. Unfortunately, this does not meet our data sharing requirements. PLOS does not permit references to inaccessible data. We require that authors provide all relevant data within the paper, Supporting Information files, or in an acceptable, public repository. Please add a citation to support this phrase or upload the data that corresponds with these findings to a stable repository (such as Figshare or Dryad) and provide and URLs, DOIs, or accession numbers that may be used to access these data. Or, if the data are not a core part of the research being presented in your study, we ask that you remove the phrase that refers to these data.
12. Please include captions for your Supporting Information files at the end of your manuscript, and update any in-text citations to match accordingly. Please see our Supporting Information guidelines for more information: http://journals.plos.org/plosone/s/supporting-information.
[Note: HTML markup is below. Please do not edit.]
Reviewers' comments:
Reviewer's Responses to Questions
Comments to the Author
1. Is the manuscript technically sound, and do the data support the conclusions?
The manuscript must describe a technically sound piece of scientific research with data that supports the conclusions. Experiments must have been conducted rigorously, with appropriate controls, replication, and sample sizes. The conclusions must be drawn appropriately based on the data presented.
Reviewer #1: Yes
Reviewer #2: Partly
**********
2. Has the statistical analysis been performed appropriately and rigorously?
Reviewer #1: Yes
Reviewer #2: Yes
**********
3. Have the authors made all data underlying the findings in their manuscript fully available?
The PLOS Data policy requires authors to make all data underlying the findings described in their manuscript fully available without restriction, with rare exception (please refer to the Data Availability Statement in the manuscript PDF file). The data should be provided as part of the manuscript or its supporting information, or deposited to a public repository. For example, in addition to summary statistics, the data points behind means, medians and variance measures should be available. If there are restrictions on publicly sharing data—e.g. participant privacy or use of data from a third party—those must be specified.
Reviewer #1: Yes
Reviewer #2: Yes
**********
4. Is the manuscript presented in an intelligible fashion and written in standard English?
PLOS ONE does not copyedit accepted manuscripts, so the language in submitted articles must be clear, correct, and unambiguous. Any typographical or grammatical errors should be corrected at revision, so please note any specific errors here.
Reviewer #1: Yes
Reviewer #2: Yes
**********
5. Review Comments to the Author
Please use the space provided to explain your answers to the questions above. You may also include additional comments for the author, including concerns about dual publication, research ethics, or publication ethics. (Please upload your review as an attachment if it exceeds 20,000 characters)
Reviewer #1: In this manuscript, the authors observed that reduced Brd2 expression benefits for extending lifespan and healthspan of C57B6/J mice. Despite Brd2 haploinsufficient mice have been generated by several groups in different ways, the pro-longevity phenotype in HET mice has never been identified. The authors also concerned the downstream effectors, e.g. Sirt1, HO-1, and P53, were upregulated in Brd2 HET tissues. These genes are apparently cytoprotective and longevity-related, partially explaining the pronounced role of Brd2 haploinsufficiency in longevity. However, there are a number of issues the authors need to address before publication.
In the second paragraph, the authors introduced Cisd2, a well-known longevity gene, prolongs the lifespan of mice to the extent that Brd2 haploinsufficiency does. Does the authors aim to exclude the benefits of Brd2 HET from Cisd2’s role? Are there some functional overlaps between these two proteins? However, I could not find any test on Cisd2 function in Brd2 HET mice.
Since the authors’ observation is so distinct to the other groups based on their mouse models, please provide the generating strategy and characterization of the Brd2 HET mice, e.g. scheme of gene trap and Brd2 protein expression profile in various tissues.
Again please discuss a bit more the reason why your mouse models are phenotypically different to other groups.
In supplementary Fig2, the author showed the bodyweight was unchanged in Brd2 HET mice when comparing to WT. However, it has been reported that Brd2 disruption in mice causes severe obesity. Please discus this difference.
In addition to kidney, the authors also definitely examined the pathologies in other organs, like liver, spleen and testis (data not shown). I suggest the authors provide these data to strengthen the conclusion that Brd2 HET improves mouse healthspan.
In this manuscript, the authors provided large descriptive information on mouse aging phenotype, yet no evidence. For example, in line 262-271, the whole paragraph demonstrated the observation of symptoms of aging in WT and HET mice without any data. The authors should provide some scientific evidence, like the pictures of aged WT and HET mice, quantifications of mouse activities, or the movies on mouse behavior …
Please label the medium lifespan of mice in figure 1. In Result section, there is no data description of Figure 2C.
Reviewer #2: This paper describes a novel discovery, that reduced expression of Brd2 leads to increased lifespan, which is associated with reduced pathology and improved health. Although this is an important observation, I have the following major concerns with the manuscript:
1. The increase in lifespan is impressive and is the key to this paper. However, the lifespan reported for the WT female and male mice (which are in the C57BL/6 background) of 23 and 26 months is very short compared to other reports (e.g., see data from Jackson Laboratory, de Cabo, and Richardson) for C57BL/6 mice. The lifespans the authors obtain with the Brd2 HET mice are actually about what is observed with normal WT mice. This is a major concern because the increased lifespan could arise from making the mice more robust to the conditions that gave rise to the shorter lifespan of the WT mice rather than retarding aging. This has been shown in previous studies, which reported increased lifespan when the median/mean lifespan of the WT mice was less thatn 26 months of age, and the increased lifespan was not replicated when conducted in colonies where the WT mice had a median/mean lifespan of over 29 months.
2. A great deal of emphasis was placed on the pathological data. The major cause of death in C57BL/6 mice is lymphoma in most aging colonies; however, the data on cancer was lumped together as tumors, i.e., it was not clear what the neoplastic lesions were in the mice. In addition, renal pathology appears to be a major problem in the mice in this study, which is in contrast to most other studies on aging in C57BL/6 mice, where renal pathology is relatively minor.
3. The data on the mice being ‘biologically’ younger was weak. For example, a great deal of emphasis was placed on the epigenetic clock. However, the epigenetic clock is a measure of chronological age, not physiological age. In addition, these data were generated with only 4 mice per group and no data were given of the animal to animal variation in data (e.g., SD or SEM). The data on observational health of the animals on page 13 was unconvincing. Data on physiological functions such as grip strength, activity, rotarod performance, and cognition would have been stronger evidence that the HET mice were physiologically younger.
Minor Concerns:
1. The authors state on page 4 (line 78-77) that there are “only a handful of candidate longevity genes for mice.” This is not correct; there have been at least two dozen different genes identified. The most cited are those that show reduced growth hormone/IGF. The Cisd2 mouse has not been studied greatly from an aging context.
2. In the methods, it was stated that the mice were either allowed to live out their lifespan or euthanized. It would be important to know the number (%) of mice euthanized for the WT and HET mice, i.e., where more WT mice euthanized.
3. Supplement: there is no list of references for the figures in the supplement, and it is not clear what the third figure is all about.
4. The introduction reads more like a discussion. For example, I found very little information about the Brd2 gene in the introduction or anywhere in the manuscript. I would have liked to know what protein this gene codes for and its biochemical/molecular function, etc.
**********
6. PLOS authors have the option to publish the peer review history of their article (what does this mean?). If published, this will include your full peer review and any attached files.
If you choose “no”, your identity will remain anonymous but your review may still be made public.
Do you want your identity to be public for this peer review? For information about this choice, including consent withdrawal, please see our Privacy Policy.
Reviewer #1: No
Reviewer #2: No
[NOTE: If reviewer comments were submitted as an attachment file, they will be attached to this email and accessible via the submission site. Please log into your account, locate the manuscript record, and check for the action link "View Attachments". If this link does not appear, there are no attachment files to be viewed.]
While revising your submission, please upload your figure files to the Preflight Analysis and Conversion Engine (PACE) digital diagnostic tool, https://pacev2.apexcovantage.com/. PACE helps ensure that figures meet PLOS requirements. To use PACE, you must first register as a user. Registration is free. Then, login and navigate to the UPLOAD tab, where you will find detailed instructions on how to use the tool. If you encounter any issues or have any questions when using PACE, please email us at gro.solp@serugif. Please note that Supporting Information files do not need this step.
- PLoS One. 2020; 15(6): e0234910. »
- Author response to Decision Letter 0
Author response to Decision Letter 0
8 May 2020
Reviewer # 1
In this manuscript, the authors observed that reduced Brd2 expression benefits for extending lifespan and healthspan of C57B6/J mice. Despite Brd2 haploinsufficient mice have been generated by several groups in different ways, the pro-longevity phenotype in HET mice has never been identified. The authors also concerned the downstream effectors, e.g. Sirt1, HO-1, and P53, were upregulated in Brd2 HET tissues. These genes are apparently cytoprotective and longevity-related, partially explaining the pronounced role of Brd2 haploinsufficiency in longevity. However, there are a number of issues the authors need to address before publication.
Comment 1: In the second paragraph, the authors introduced Cisd2, a well-known longevity gene, prolongs the lifespan of mice to the extent that Brd2 haploinsufficiency does. Does the authors aim to exclude the benefits of Brd2 HET from Cisd2’s role? Are there some functional overlaps between these two proteins? However, I could not find any test on Cisd2 function in Brd2 HET mice.
Response 1: First, our HET mice are identical to C57B6/J at every gene except Brd2, having been back-crossed for at least 10 generations. Thus, there is little or no probability that any meaningful Cisd2 expression differences between WTs and HETs exist. Second, there are no known functional overlaps between Cisd2 (which is chiefly involved in Calcium homeostasis) and Brd2 (which is an epigenetic reader). These points have now been included in the second paragraph of Introduction and Discussion (lines 430-435).
Comment 2: Since the authors’ observation is so distinct to the other groups based on their mouse models, please provide the generating strategy and characterization of the Brd2 HET mice, e.g. scheme of gene trap and Brd2 protein expression profile in various tissues.
Response 2: Respectfully, we must disagree with the Reviewer’s comment that our results are “so distinct” from other groups. First, Gyruis et al. (2009) (1) used the same ES cell line as we did (RRE050) and confirmed our findings (Shang et al. 2009) (2), including that Brd2-/- mice have impaired CNS development, an observation about which the Wang et al. (2009) (3) paper is silent. Second, Wang et al. used a Brd2 insertion/disrupter (e.g. ES cell line RRT234) different from the one we used or that Gyruis et al used. Third, Wang et al’s principal phenotype was obesity in the mice but neither Gyruis et al nor we observed obesity in our mice. Fourth, and most quizzically, there is residual Brd2 expression in their double knockout mouse, whereas neither Gyruis nor we observed any residual expression. Thus, the phenotype of Gyruis’s mouse and our mouse phenotype are not “distinct” whereas the Wang et al. phenotype differs markedly from our findings and those of Gyruis.
As to generating strategy, the genetic engineering and characteristics of Brd2 HET mice has been described in details in Shang et al (2009) (2), and we have added the mouse generation strategy in detail in the revised manuscript (see Materials and Methods, lines 143-151).
Gene expression profiles showing (among other things) the reduction of Brd2 expression in HET animals in kidney, liver, heart and mouse embryonic fibroblasts is a part of supplemental information (Supplementary Figure 1). These findings were also replicated by Gyruis et al. (2009)(1).
Comment 3: Again, please discuss a bit more the reason why your mouse models are phenotypically different to other groups.
Response 3: This comment was addressed above in Response 2 and we have modified our discussion incorporating the genotypic and phenotypic differences related to the different mouse models (Discussion, lines 437-444).
Comment 4: In supplementary Fig2, the author showed the bodyweight was unchanged in Brd2 HET mice when comparing to WT. However, it has been reported that Brd2 disruption in mice causes severe obesity. Please discus this difference.
Response 4: Again, this comment was addressed in Response 2, and in Discussion.
Comment 5: In addition to kidney, the authors also definitely examined the pathologies in other organs, like liver, spleen and testis (data not shown). I suggest the authors provide these data to strengthen the conclusion that Brd2 HET improves mouse health span.
Response 5: The histopathology data for liver, spleen and testis has been included in the supplementary information in the revised manuscript.
Comment 6: In this manuscript, the authors provided large descriptive information on mouse aging phenotype, yet no evidence.. For example, in line 262-271, the whole paragraph demonstrated the observation of symptoms of aging in WT and HET mice without any data. The authors should provide some scientific evidence, like the pictures of aged WT and HET mice, quantifications of mouse activities, or the movies on mouse behavior …
Response 6: We are somewhat surprised by the comment about the acceptability our description of the differences over time between the HET and WT mice. Such descriptive content has long been a staple of scientific publications going back to the very beginning of science journals. Not surprisingly, standard photographs are often inadequate to capture the differences (e.g. subtle differences in grooming and grizzled appearance). Furthermore, while lifetime surveillance of each mouse would permit quantification of differences in maintenance of youthful activity versus continued decline in actively resulting in lethargy, this is impractical. Therefore, the time-honored practice of reporting such observations in the scientific literature are crucial to the understanding of phenotype.
As to mouse behavior, those results were published in Chachua et al. 2014 (4). Those studies showed that while HETs and WTs do have similar cognitive functioning, the HETs have decreased anxiety, and are more aggressive than WTs (especially in the case of females).
Comment 7: Please label the medium lifespan of mice in figure 1. In Result section, there is no data description of Figure 2C.
Response 7: The median lifespan has been labeled in Figure 1, and the Figure 2C has been mentioned in the revised manuscript (Line 350).
Reviewer # 2
This paper describes a novel discovery, that reduced expression of Brd2 leads to increased lifespan, which is associated with reduced pathology and improved health. Although this is an important observation, I have the following major concerns with the manuscript:
Comment 1A: The increase in lifespan is impressive and is the key to this paper. However, the lifespan reported for the WT female and male mice (which are in the C57BL/6 background) of 23 and 26 months is very short compared to other reports (e.g., see data from Jackson Laboratory, de Cabo, and Richardson) for C57BL/6 mice.
Response 1A: In fact, there is no appreciable difference in the median lifespans of our WT males and the Jackson Lab (JAX) C57BL/6J mouse colony. In our longevity tabulations we included all mice that died before adulthood, which Jackson labs does not. Furthermore, there is notable variations in average lifespan of this strain of mice across vivaria, especially differences in deciding when and with what pathologies aging mice should be euthanized (5, 6). Furthermore, although the salient comparison is the one between our HETs and our WTs, it is also true that our HETs live longer (on average) than Jackson Labs C57BL/6J WT mice (p<0.002) (7). This is included in Discussion section in the revised manuscript. (See also the answer to 1C, below.)
Comment 1B: The lifespans the authors obtain with the Brd2 HET mice are actually about what is observed with normal WT mice.
Response 1B: We think the reviewer is mistaken on this point. 31.5 months (HET median) is 23% longer than 26.5 months (WT median; p<0.0001).
Comment 1C: This is a major concern because the increased lifespan could arise from making the mice more robust to the conditions that gave rise to the shorter lifespan of the WT mice rather than retarding aging. This has been shown in previous studies, which reported increased lifespan when the median/mean lifespan of the WT mice was less than 26 months of age, and the increased lifespan was not replicated when conducted in colonies where the WT mice had a median/mean lifespan of over 29 months.
Response 1C: There are several reports in the literature of median lifespan for WT approx. 26-27 months, so by comparison, our HET animal with median lifespan of 31-32 months is still quite remarkable. The HET males showed a 19% extension in median lifespan (925 vs 781 days; p<0.0001) but the WT females appear to have shorter lives than the WT males: WT females show a median lifespan of 23 months, and we agree it would be interesting to know how Brd2 haploinsufficiency might compensate for it compared to 26 months for WT males, but we hypothesize that the reason for the difference is not genetic. The median lifespan of our WT males is the same as reported by other vivaria, but the mean figure for WT females is lower than the median lifespan of JAX C57. We hypothesize that the hyper-aggressiveness of the HET females (4), which are more aggressive than even the WT males, leads to increased stress on the WT females, which are housed with the HET females, and that this accounts for the somewhat shorter lifespan of the WT females, which also reduces the overall WT survival rate. We plan to test this hypothesis but currently, such investigations are beyond the scope of the current paper. Moreover, it is highly unlikely that the shortened lifespan of WT females is genetic in origin since our HETs (which were backcrossed for 10 generations with WTs) do not have hidden mutations or deletions (as determined by whole genome sequencing).
Comment 2: A great deal of emphasis was placed on the pathological data. The major cause of death in C57BL/6 mice is lymphoma in most aging colonies; however, the data on cancer was lumped together as tumors, i.e., it was not clear what the neoplastic lesions were in the mice. In addition, renal pathology appears to be a major problem in the mice in this study, which is in contrast to most other studies on aging in C57BL/6 mice, where renal pathology is relatively minor.
Response 2: Our lab has considerable expertise in the area of kidney and liver pathology, and there is strong evidence in the aging literature (8, 9) that dysmorphic kidney and liver structure (e.g. lesions) is a normal part of the aging process in mice. Therefore, differences in the organ pathology of HET and WT mice are a natural place to start: both from the perspective of documenting what was observed, and from the perspective of generating mechanistic hypotheses to test. For instance, given the abnormal structure of both liver and kidney in WT (Supplementary Figure 3), it is possible that HET animals may have fewer senescent cells than WT. This would be an interesting hypothesis to test in future studies. The overwhelming point, however, is that the HETs show considerably less age-related organ pathology than the WT.
While lymphoma is most certainly an important cause of death in C57 (somewhere between 10% and 50% (10), our primary endpoint for this study is lifespan, not cancer. It was only incidentally (ie, at the time of death), that we noticed a difference in HETs and WTs with respect to the presence of solid tumor(s). While determination of the primary tumor type in these animals would be interesting, this is beyond the scope of this current work.
Comment 3A: The data on the mice being ‘biologically’ younger was weak. For example, a great deal of emphasis was placed on the epigenetic clock. However, the epigenetic clock is a measure of chronological age, not physiological age.
Response 3A: The methylation clock (DNAge) was first demonstrated to be highly correlated with chronological age, and could then be used as a good predictor of biological age (11-15).
Furthermore, as we illustrate in Table 1, WT mice of the same chronological age as HET mice (i.e., 770 days) have dramatically higher biologically measured age (DNAges) than the longer-lived HET mice. Since we know that HETs live longer than WTs (in general), this confirms several reports in the aging literature that DNAge is a better measure of biological age than is simple chronological age.
Comment 3B: In addition, these data were generated with only 4 mice per group and no data were given of the animal to animal variation in data (e.g., SD or SEM).
Response 3B: Even though there are only 4 mice per group, the methylation clock analysis showed statistically significant younger liver tissue in HET’s than WT’s (p<0.012 and SEM(WT): 4.5 and SEM(HET): 7.1), and the DNAges of each animal are shown in Table 2. We have included SEMs in the revised manuscript.
Comment 3C: The data on observational health of the animals on page 13 was unconvincing.
Response 3C: Please see Response 6 to Reviewer 1.
Comment 3D: Data on physiological functions such as grip strength, activity, rotarod performance, and cognition would have been stronger evidence that the HET mice were physiologically younger.
Response 3D: We have previously tested HET and WT mice in a battery of behavioral tests (open field, tube dominance test, elevated plus maze, Morris water maze and Barnes maze) that showed increased aggressiveness in HETs without cognitive impairment (4). Relative to the biological indicators of age-related differences that we already have described (e.g. HETs have increased lifespan, HETs have reduced cancer incidence, and HET have reduced organ pathology), tracking the rate of cognitive decline will not give us a better indication of the HET-WT aging difference. Testing such physiological functions in HET mice is currently beyond the scope of this manuscript, and is contingent upon future funding.
Minor Concerns:
Comment 4: The authors state on page 4 (line 78-77) that there are “only a handful of candidate longevity genes for mice.” This is not correct; there have been at least two dozen different genes identified. The most cited are those that show reduced growth hormone/IGF. The Cisd2 mouse has not been studied greatly from an aging context.
Response 4: By our count, we find that there are only 11 candidate longevity genes for C57 mice (16), and we now express this explicitly in the revised manuscript in the second paragraph of Introduction. Moreover, of the aforementioned 11 candidates, only one has greater median lifespan than our HETs, and two actually have reduced lifespan compared to JAX labs.
Comment 5: In the methods, it was stated that the mice were either allowed to live out their lifespan or euthanized. It would be important to know the number (%) of mice euthanized for the WT and HET mice, i.e., where more WT mice euthanized.
Response 5: In the WT cohort, 68% mice were euthanized as opposed to 45% mice in HET group.
Comment 6: Supplement: there is no list of references for the figures in the supplement, and it is not clear what the third figure is all about.
Response 6: Figures in the Supplementary information has been appropriately cited in the manuscript and the third figure in the supplementary information has been deleted.
Comment 7: The introduction reads more like a discussion. For example, I found very little information about the Brd2 gene in the introduction or anywhere in the manuscript. I would have liked to know what protein this gene codes for and its biochemical/molecular function, etc.
Response 7: The reviewer raises an important stylistic point. We did not include much information about Brd2’s biochemical/molecular function because, as interesting as that would be from a basic science point of view, we have yet to find a plausible explanation for the wide range of biological processes in which this molecule is involved. Aside from its two most cited roles: a transcription factor element in copying RNA from DNA, and an acetylated histone effector, it is associated with a number of disparate biologic observations, such as: over-expression in cancer; regulation of neuron migration during development; linkage and association to a specific form of epilepsy; over-expression in hormonally modulated epithelia (e.g., mammary gland, ovary, kidney, and uterus); and it is even reported to act as a scaffold for TATA binding. Thus, in our view, what the protein “does” is not at all well understood. Furthermore, because it takes a great deal of space to explain what individual processes the protein is involved in, and because this explanation is unlikely to provide an overarching understanding of how Brd2 brings about these observations, we did not feel it at all appropriate to go into detail about topics that do not, as far as we can see, increase understanding of Brd2’s effect on mouse lifespan and health. Instead, we opted for explaining only how we came to serendipitously observe its effect on longevity and to substantiate the depth of that effect with observations, for example, on changes in kidney pathology between HETs and WTs. In truth, we regret not being able to review Brd2’s biologic functions in greater detail than the few lines allotted in Discussion, but that task is best left to a review article, not in this report—which emphasizes Brd2’s dramatic and surprisingly beneficial effect on mouse longevity and health.
References:
1. Gyuris A, Donovan DJ, Seymour KA, Lovasco LA, Smilowitz NR, Halperin AL, et al. The chromatin-targeting protein Brd2 is required for neural tube closure and embryogenesis. Biochim Biophys Acta. 2009;1789(5):413-21.
2. Shang E, Wang X, Wen D, Greenberg DA, Wolgemuth DJ. Double bromodomain-containing gene Brd2 is essential for embryonic development in mouse. Dev Dyn. 2009;238(4):908-17.
3. Wang F, Liu H, Blanton WP, Belkina A, Lebrasseur NK, Denis GV. Brd2 disruption in mice causes severe obesity without Type 2 diabetes. Biochem J. 2009;425(1):71-83.
4. Chachua T, Goletiani C, Maglakelidze G, Sidyelyeva G, Daniel M, Morris E, et al. Sex-specific behavioral traits in the Brd2 mouse model of juvenile myoclonic epilepsy. Genes Brain Behav. 2014;13(7):702-12.
5. Yuan R, Peters LL, Paigen B. Mice as a mammalian model for research on the genetics of aging. ILAR J. 2011;52(1):4-15.
6. Yuan R, Tsaih SW, Petkova SB, Marin de Evsikova C, Xing S, Marion MA, et al. Aging in inbred strains of mice: study design and interim report on median lifespans and circulating IGF1 levels. Aging Cell. 2009;8(3):277-87.
7. Flurkey K. The Mouse in Biomedical Research. 2nd Edition ed. Burlington, MA: Elsevier; 2007.
8. Lim JH, Kim EN, Kim MY, Chung S, Shin SJ, Kim HW, et al. Age-associated molecular changes in the kidney in aged mice. Oxid Med Cell Longev. 2012;2012:171383.
9. Pettan-Brewer C, Treuting PM. Practical pathology of aging mice. Pathobiol Aging Age Relat Dis. 2011;1.
10. Ward JM. Lymphomas and leukemias in mice. Exp Toxicol Pathol. 2006;57(5-6):377-81.
11. Bocklandt S, Lin W, Sehl ME, Sanchez FJ, Sinsheimer JS, Horvath S, et al. Epigenetic predictor of age. PLoS One. 2011;6(6):e14821.
12. Zhang Y, Wilson R, Heiss J, Breitling LP, Saum KU, Schottker B, et al. DNA methylation signatures in peripheral blood strongly predict all-cause mortality. Nat Commun. 2017;8:14617.
13. Petkovich DA, Podolskiy DI, Lobanov AV, Lee SG, Miller RA, Gladyshev VN. Using DNA Methylation Profiling to Evaluate Biological Age and Longevity Interventions. Cell Metab. 2017;25(4):954-60 e6.
14. Field AE, Robertson NA, Wang T, Havas A, Ideker T, Adams PD. DNA Methylation Clocks in Aging: Categories, Causes, and Consequences. Mol Cell. 2018;71(6):882-95.
15. Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14(10):R115.
16. Pedro de Magalhaes J, Thompson L, de Lima I, Gaskill D, Li X, Thornton D, et al. A Reassessment of Genes Modulating Aging in Mice Using Demographic Measurements of the Rate of Aging. Genetics. 2018;208(4):1617-30.
Attachment
Submitted filename: Response to Reviewers.docx
- PLoS One. 2020; 15(6): e0234910. »
- Decision Letter 1
Decision Letter 1
5 Jun 2020
Brd2 haploinsufficiency extends lifespan and healthspan in C57B6/J mice
PONE-D-19-32190R1
Dear Dr. Pathak,
We’re pleased to inform you that your manuscript has been judged scientifically suitable for publication and will be formally accepted for publication once it meets all outstanding technical requirements.
Within one week, you’ll receive an e-mail detailing the required amendments. When these have been addressed, you’ll receive a formal acceptance letter and your manuscript will be scheduled for publication.
An invoice for payment will follow shortly after the formal acceptance. To ensure an efficient process, please log into Editorial Manager at http://www.editorialmanager.com/pone/, click the 'Update My Information' link at the top of the page, and double check that your user information is up-to-date. If you have any billing related questions, please contact our Author Billing department directly at gro.solp@gnillibrohtua.
If your institution or institutions have a press office, please notify them about your upcoming paper to help maximize its impact. If they’ll be preparing press materials, please inform our press team as soon as possible -- no later than 48 hours after receiving the formal acceptance. Your manuscript will remain under strict press embargo until 2 pm Eastern Time on the date of publication. For more information, please contact gro.solp@sserpeno.
Kind regards,
Christoph Englert
Academic Editor
PLOS ONE
Additional Editor Comments (optional):
Reviewers' comments:
Reviewer's Responses to Questions
Comments to the Author
1. If the authors have adequately addressed your comments raised in a previous round of review and you feel that this manuscript is now acceptable for publication, you may indicate that here to bypass the “Comments to the Author” section, enter your conflict of interest statement in the “Confidential to Editor” section, and submit your "Accept" recommendation.
Reviewer #1: All comments have been addressed
**********
2. Is the manuscript technically sound, and do the data support the conclusions?
The manuscript must describe a technically sound piece of scientific research with data that supports the conclusions. Experiments must have been conducted rigorously, with appropriate controls, replication, and sample sizes. The conclusions must be drawn appropriately based on the data presented.
Reviewer #1: (No Response)
**********
3. Has the statistical analysis been performed appropriately and rigorously?
Reviewer #1: (No Response)
**********
4. Have the authors made all data underlying the findings in their manuscript fully available?
The PLOS Data policy requires authors to make all data underlying the findings described in their manuscript fully available without restriction, with rare exception (please refer to the Data Availability Statement in the manuscript PDF file). The data should be provided as part of the manuscript or its supporting information, or deposited to a public repository. For example, in addition to summary statistics, the data points behind means, medians and variance measures should be available. If there are restrictions on publicly sharing data—e.g. participant privacy or use of data from a third party—those must be specified.
Reviewer #1: (No Response)
**********
5. Is the manuscript presented in an intelligible fashion and written in standard English?
PLOS ONE does not copyedit accepted manuscripts, so the language in submitted articles must be clear, correct, and unambiguous. Any typographical or grammatical errors should be corrected at revision, so please note any specific errors here.
Reviewer #1: (No Response)
**********
6. Review Comments to the Author
Please use the space provided to explain your answers to the questions above. You may also include additional comments for the author, including concerns about dual publication, research ethics, or publication ethics. (Please upload your review as an attachment if it exceeds 20,000 characters)
Reviewer #1: (No Response)
**********
7. PLOS authors have the option to publish the peer review history of their article (what does this mean?). If published, this will include your full peer review and any attached files.
If you choose “no”, your identity will remain anonymous but your review may still be made public.
Do you want your identity to be public for this peer review? For information about this choice, including consent withdrawal, please see our Privacy Policy.
Reviewer #1: No
- PLoS One. 2020; 15(6): e0234910. »
- Acceptance letter
Acceptance letter
10 Jun 2020
PONE-D-19-32190R1
Brd2 haploinsufficiency extends lifespan and healthspan in C57B6/J mice
Dear Dr. Pathak:
I'm pleased to inform you that your manuscript has been deemed suitable for publication in PLOS ONE. Congratulations! Your manuscript is now with our production department.
If your institution or institutions have a press office, please let them know about your upcoming paper now to help maximize its impact. If they'll be preparing press materials, please inform our press team within the next 48 hours. Your manuscript will remain under strict press embargo until 2 pm Eastern Time on the date of publication. For more information please contact gro.solp@sserpeno.
If we can help with anything else, please email us at gro.solp@enosolp.
Thank you for submitting your work to PLOS ONE and supporting open access.
Kind regards,
PLOS ONE Editorial Office Staff
on behalf of
Dr. Christoph Englert
Academic Editor
PLOS ONE