- Journal List
- Genetics
- v.208(4); 2018 Apr
- PMC5887152

A Reassessment of Genes Modulating Aging in Mice Using Demographic Measurements of the Rate of Aging
Associated Data
- Supplementary Materials
- 1617FileS1.xlsx (136K)GUID: FD7F9C8D-6B4A-4738-8765-321B4E2DC5FB1617FileS2.txt (4.1K)GUID: FDABB2DF-B676-4C0C-B882-7A1110DCF14F
- Data Availability Statement
The survival data used in this study is provided in the Supplemental Material, Tables S1 and S2 in File S1. The SPSS code used is provided in File S2.
Abstract
Many studies have reported genetic interventions that have an effect on mouse life span; however, it is crucial to discriminate between manipulations of aging and aging-independent causes of life extension. Here, we used the Gompertz equation to determine whether previously reported aging-related mouse genes statistically affect the demographic rate of aging. Of 30 genetic manipulations previously reported to extend life span, for only two we found evidence of retarding demographic aging: Cisd2 and hMTH1. Of 24 genetic manipulations reported to shorten life span and induce premature aging features, we found evidence of five accelerating demographic aging: Casp2, Fn1, IKK-β, JunD, and Stub1. Overall, our reassessment found that only 15% of the genetic manipulations analyzed significantly affected the demographic rate of aging as predicted, suggesting that a relatively small proportion of interventions affecting longevity do so by regulating the rate of aging. By contrast, genetic manipulations affecting longevity tend to impact on aging-independent mortality. Our meta-analysis of multiple mouse longevity studies also reveals substantial variation in the controls used across experiments, suggesting that a short life span of controls is a potential source of bias. Overall, the present work leads to a reassessment of genes affecting the aging process in mice, with broad implications for our understanding of the genetics of mammalian aging and which genes may be more promising targets for drug discovery.
THE discovery that single-gene manipulations can significantly modulate longevity is arguably the major breakthrough in biogerontology thus far (Kenyon 2010). Genetic manipulations of aging in mice are crucial to gather insights into the underlying mechanisms of aging (de Magalhães and Faragher 2008), to discover pathways modulating longevity (Fernandes et al. 2016), and to identify candidate genes for drug discovery (de Magalhães et al. 2012; Barardo et al. 2017). Moreover, the manipulation of the aging process in mammalian models (particularly mice) via genetic manipulation (gene knockouts, over expression, etc.) is crucial to test mechanistic hypotheses of aging (de Magalhães 2005). However, determining if such genetic interventions actually affect the aging process and not some other factor(s) of health is not always straight forward. For example, should a genetic intervention reduce an organism’s resistance to disease, this could conceivably reduce the life span of the organism, although the rate of aging would not have been affected. Differentiating between genetic interventions that affect the life span of an organism through altered health as opposed to changes in the rate of aging is therefore essential to gain insights on aging and determine interventions with wide-ranging effects (Hayflick 2000; de Magalhães et al. 2005).
There are two fundamental methods to determine if a life-extending genetic intervention has altered the rate of aging rather than general health. One can track the onset and progression of age-related ailments and physiological degeneration to determine if there is a shift in the onset and progression of the ailments. In addition, efforts have been made to quantify aging rates with mathematical models. The Gompertz law of mortality:
where Rm(t) is the hazard or mortality rate (likelihood of death at any time), t is age, R0 is the age-independent mortality component, and α is the age-specific exponential coefficient, describes how the hazard rate increases exponentially with age (Finch and Pike 1996). From the Gompertz parameters, the mortality rate doubling time (MRDT) can be calculated. As defined by Finch (1990), the MRDT is the amount of time it takes for the mortality rate to double for a given cohort. A change in MRDT indicates a change in the demographic rate of aging, which is not a perfect reflection of biological aging but a metric that correlates with physiological deterioration and health (Finch 1990; de Magalhães et al. 2005). Although some mouse studies have investigated MRDT (Hinkal et al. 2009; Lapointe et al. 2009), many authors still often assume that changes in the life span of mice following a genetic intervention directly equate to changes in the rate of aging, leading to the misrepresentation of certain genes as having a causal role in aging, when in reality they do not.
Many studies have reported altered median and/or maximum life span as a result of an intervention, but life span alterations may have a number of causes, including altered age at onset of senescence and age-independent mortality (Pletcher et al. 2000). To address this lack of distinction, we previously used linear regression to fit the Gompertz model to longevity data from published mouse studies, and statistically compared the rates of aging in these cohorts (de Magalhães et al. 2005). For example, we showed that caloric restriction increases the MRDT and thus retards the demographic rate of aging (de Magalhães et al. 2005). Here, the same methodology was employed to reassess mouse longevity data published since 2005 and to identify which genes are more important in determining the demographic rate of aging. Lastly, we perform a meta-analysis combining the data from the present study and from our 2005 analysis to investigate patterns in how longevity changes correlate with changes in demographic rates of aging.
Methods
Data selection and extraction
Studies published since 2005 were selected since studies published up to 2005 were analyzed previously (de Magalhães et al. 2005). Genes were selected from the GenAge database, build 17 (Tacutu et al. 2013). GenAge already excludes genes extending life span in short-lived (or disease) mutants or conditions. In addition, some genes were excluded as they could not be studied for demographic parameters (some genes could not be properly fitted to the Gompertz model and some studies lacked full life span data, while other studies lacked sufficient cohort size). A total of 54 genetic manipulations could be properly analyzed (primarily single-gene manipulations but also some manipulations involving more than one gene): 30 genetic manipulations that extended mouse life span (Table 1) and 24 genetic manipulations that reduced life span (Table 2).
Table 1
| Gene or protein | Type | Strain | Gender | n | t50 (yr) | tmax (yr) | Reference |
|---|---|---|---|---|---|---|---|
| Adcy5 | WT | 129/SvJ-C57BL/6 | M & F | 25 | 2.08 | 2.75 | Yan et al. (2007) |
| −/− | M & F | 13 | 2.75 | 3.08 | |||
| Agrt1a | WT | C57BL/6 × 129/SvEv | M | 10 | 2.08 | 2.41 | Benigni et al. (2009) |
| −/− | M | 20 | 2.63 | 3.00 | |||
| Arf/p53 | WT | C57BL/6J | 111 | 2.31 | 3.30 | Matheu et al. (2007) | |
| s-Arf/p53 | 25 | 2.63 | 3.03 | ||||
| Atg5 | WT | C57BL/6 | 65 | 1.93 | 2.14 | Pyo et al. (2013) | |
| Atg5-Tg | 70 | 2.26 | 2.52 | ||||
| BubR1 | WT | C57BL/6-SV129 | M:30 | 60 | 1.72 | 2.9 | Baker et al. (2013) |
| F: 30 | |||||||
| mBubR1-Tg | M:29 | 57 | 2.02 | 3.42 | |||
| F:28 | |||||||
| Cat | WT | 4033 | M & F | 44 | 2.16 | 2.83 | Schriner et al. (2005) |
| MCAT | 4033 | M & F | 20 | 2.58 | 3.10 | ||
| WT | 4403 | M & F | 58 | 2.18 | 3.06 | ||
| MCAT | 4403 | M & F | 42 | 2.60 | 3.33 | ||
| Cisd2 | WT (M) | C57BL/6 | M | 40 | 2.25 | 2.90 | Wu et al. (2012) |
| Cisd2-Tg (M) | M | 34 | 2.69 | 3.07 | |||
| WT (F) | F | 25 | 2.27 | 2.50 | |||
| Cisd2-Tg (F) | F | 21 | 2.74 | 3.25 | |||
| Clk-1 | WT | 129Sv/J | F | 12 | 2.01 | 2.30 | Liu et al. (2005) |
| +/− | F | 10 | 2.34 | 2.54 | |||
| Dgat1 | WT | C57BL/6J | F | 30 | 2.04 | 2.81 | Streeper et al. (2012) |
| −/− | F | 30 | 2.58 | 3.05 | |||
| Esp8 | WT | C57BL/6 | M:13 | 29 | 1.74 | 2.72 | Tocchetti et al. (2010) |
| F:16 | |||||||
| −/− | M:20 | 39 | 2.16 | 3.00 | |||
| F:19 | |||||||
| FGF21 | WT | C57Bl/6J | M:32 | 67 | 2.34 | 3.56 | Zhang et al. (2012) |
| F:35 | |||||||
| FGF21-Tg | M:37 | 77 | 3.18 | NR (> 3.65) | |||
| F:40 | |||||||
| Ghrh | WT | C57BL/6 | M:56 | 108 | 1.75 | 3.20 | Sun et al. (2013) |
| × 129SV | F:52 | ||||||
| KO | M:39 | 97 | 2.55 | 3.58 | |||
| F:58 | |||||||
| Gpx4 | WT | C57BL/6 | 50 | 2.63 | 3.34 | Ran et al. (2007) | |
| +/– | 50 | 2.83 | 3.17 | ||||
| hMTH1 | WT | C57BL/6 | M | 42 | 2.17 | 2.64 | De Luca et al. (2013) |
| hMTH1-Tg | M | 34 | 2.51 | 3.21 | |||
| Htt | WT | 129/Sv- | M:1 | 15 | 2.34 | 2.93 | Zheng et al. (2010) |
| F:14 | |||||||
| ΔQ/ΔQ | M:2 | 15 | 2.76 | 3.43 | |||
| F:13 | |||||||
| Igf1 | WT | FVB | M | 39 | 1.97 | 3.17 | Li and Ren (2007) |
| Igf1-Tg | M | 38 | 2.43 | 3.42 | |||
| IκB-α | MBH-GFP | C57BL/6 | 23 | 2.41 | 2.91 | Zhang et al. (2013) | |
| MBH-IκB-α | 31 | 2.64 | 3.09 | ||||
| Irs1 | WT (F) | C57BL/6 | F | 21 | 2.02 | 2.86 | Selman et al. (2009) |
| −/− (F) | F | 14 | 2.66 | 3.61 | |||
| Irs2 | WT | C57BL/6J | M:21 | 93 | 2.36 | 2.86 | Taguchi et al. (2007) |
| F:30 | |||||||
| Brain-specific +/− | M:27 | 65 | 2.81 | 3.34 | |||
| F:60 | |||||||
| Mif | WT | C57BL/6J × 129/SvJ | F | 24 | 2.01 | 2.70 | Harper et al. (2010) |
| −/− | F | 39 | 2.45 | 3.51 | |||
| mTOR | WT | 129 × C57BL/6 | M & F | 34 | 2.13 | 3.14 | Wu et al. (2013) |
| KO | M & F | 43 | 2.51 | 3.14 | |||
| Myc | WT (F) | C57BL/6 | F | 37 | 2.23 | 2.86 | Hofmann et al. (2015) |
| +/− (F) | F | 39 | 2.68 | 3.58 | |||
| WT (M) | M | 42 | 2.41 | 3.00 | |||
| +/− (M) | M | 42 | 2.66 | 3.26 | |||
| PAPP-A | WT | C57BL6 × 129SV/E | M & F | 21 | 1.84 | 2.44 | Conover and Bale (2007) |
| −/− | M & F | 20 | 2.64 | 3.11 | |||
| Pten | WT | C57BL/6 | M:49 | 112 | 2.17 | 2.92 | Ortega-Molina et al. (2012) |
| × CBA | F:63 | ||||||
| Pten-Tg | M:32 | 64 | 2.44 | 3.21 | |||
| F:32 | |||||||
| RpS6K1 | WT | C57BL/6 | M:26 | 49 | 2.23 | 3.00 | Selman et al. (2009) |
| F:23 | |||||||
| −/− | M:19 | 48 | 2.64 | 3.40 | |||
| F:29 | |||||||
| RIIβ | WT | C57BL/6 (males) | M | 20 | 2.42 | 2.79 | Enns et al. (2009) |
| RIIB −/− | M | 20 | 2.75 | 3.07 | |||
| Sirt1 | WT | C57BL/6 | M & F | 31 | 2.30 | 3.01 | Satoh et al. (2013) |
| Brain-specific Tg | M & F | 34 | 2.56 | 3.11 | |||
| Slc13a1 | WT (M) | C57BL/6J × 129/SV | M | 21 | 1.93 | 2.60 | Markovich et al. (2011) |
| Nas1 −/− (M) | M | 25 | 2.54 | 3.30 | |||
| WT (F) | F | 34 | 1.68 | 2.50 | |||
| Nas1 −/− (F) | F | 38 | 2.06 | 2.90 | |||
| Surf1 | WT | BDF1 × cre | M:23 | 48 | 1.78 | NR | Dell’agnello et al. (2007) |
| F:25 | |||||||
| −/− | M:21 | 43 | 2.17 | 2.48 | |||
| F:22 | |||||||
| Tert | Sp53 | C57BL/6 × DBA/2 | M & F | 68 | 2.13 | 3.09 | Tomás-Loba et al. (2008) |
| Sp53/TgTert | M & F | 56 | 2.36 | 3.22 | |||
| Sp53/Sp16/SArf | M & F | 39 | 2.38 | 3.18 | |||
| Sp53/Sp16/SArf/TgTert | M & F | 27 | 2.38 | 3.26 |
t50, median life span; tmax, maximum life span; yr, year; WT, wild-type; M, male; F, female; Tg, transgenic; NR, not reported; KO, knockout.
Table 2
| Gene or protein | Type | Strain | Gender | n | t50 (yr) | tmax (yr) | Reference |
|---|---|---|---|---|---|---|---|
| Aag, Atm, Mgmt | WT | C57BL/6 | M & F | 37 | 2.03 | 2.87 | Meira et al. (2014) |
| Aag –/– | M & F | 29 | 1.80 | 2.50 | |||
| Mgmt –/– | M & F | 50 | 1.92 | 2.77 | |||
| Atm –/– | M & F | 19 | 0.56 | 1.37 | |||
| Aag –/– Mgmt –/– | M & F | 31 | 1.67 | 2.71 | |||
| ATR | WT | M & F | 20 | NR | NR | Murga et al. (2009) | |
| ATRs/s | M & F | 27 | 0.42 | 0.65 | |||
| Brca1 | WT | 129O1a × C57BL/6J | F | 32 | 2.15 | 2.56 | Jeng et al. (2007) |
| +/− | F | 26 | 1.94 | 2.53 | |||
| Bub3 + Rae1 | WT | 129Sv/E × C57BL/6 | N/A | 70 | 2.08 | NR | Baker et al. (2006) |
| +/−, +/− | N/A | 100 | 1.84 | NR | |||
| Casp2 | WT | C57BL/6 | N/A | 64 | 2.62 | 3.51 | Zhang et al. (2007) |
| −/− | N/A | 64 | 2.62 | 3.25 | |||
| Cdc42 | WT | C57BL/6+/− 129/Sv | M & F | 16 | 2.28 | 2.49 | Wang et al. (2007) |
| −/− | M & F | 21 | 1.09 | 2.50 | |||
| Cisd2 | WT | C57BL/6 (B6) | M & F | 49 | 2.09 | 2.53 | Chen et al. (2009) |
| Cisd2−/− | M & F | 16 | 1.28 | 2.15 | |||
| Cisd2 | WT | C57BL/6 | M | 40 | 2.25 | 2.90 | Wu et al. (2012) |
| +/– | M | 51 | 2.05 | 2.57 | |||
| –/– | M | 27 | 1.76 | 2.32 | |||
| WT | F | 25 | 2.27 | 2.50 | |||
| +/– | F | 47 | 1.92 | 2.50 | |||
| –/– | F | 49 | 1.83 | 2.94 | |||
| DNA pol β | WT | C57BL/6 | M | 60 | 2.54 | 3.19 | Cabelof et al. (2006) |
| +/– | M | 67 | 2.54 | 3.19 | |||
| Fgf-23 | −/− | Sv129J | M & F | 15 | 0.12 | 0.26 | Razzaque et al. (2006) |
| Fn1 | WT | C57BL/6 | M | 39 | 2.43 | NR | Muro et al. (2003) |
| Fn1EDA −/− | M | 53 | 1.93 | NR | |||
| HtrA2/Omi | mnd2/+;Tg | C57BL/6J | M & F | 23 | N/A | N/A | Kang et al. (2013) |
| mnd2/mnd2;Tg | M & F | 21 | 1.28 | 1.50 | |||
| Htr1b | WT | N/A | 21 | 2.58 | 3.14 | Sibille et al. (2007) | |
| −/− | N/A | 24 | 2.05 | 2.75 | |||
| IKK-β | MBH-GFP | C57BL/6 | M | 23 | 2.41 | 2.91 | Zhang et al. (2013) |
| N/Ikbkbl/l | M | 24 | 2.23 | 2.56 | |||
| junD | WT | N/A | M & F | 35 | 1.91 | 2.41 | Laurent et al. (2008) |
| JunD−/− | N/A | M & F | 35 | 1.66 | 2.17 | ||
| Msh2 | WT | 129S/SvEvTac | M & F | 51 | NR | NR | Wei et al. (2003) |
| −/− | M & F | 32 | 0.58 | 0.92 | |||
| Pasg | −/− | 129/SvJ/C57BL/6J | N/A | 63 | 0.01 | 0.07 | Sun et al. (2004) |
| Pparg | WT | F | 25 | 2.35 | 2.68 | Argmann et al. (2009) | |
| Pparg2−/− | F | 26 | 2.14 | 2.62 | |||
| Sirt7 | WT | C57Bl/6 × 129Sv | M | 98 | NR | NR | Vakhrusheva et al. (2008) |
| −/− | M | 32 | 0.74 | 1.60 | |||
| Socs2 | WT | C57BL/6J × FVB | M & F | 123 | 2.10 | 3.00 | Casellas and Medrano (2008) |
| hg/hg | M & F | 146 | 1.33 | 2.59 | |||
| Stub1 | WT | C57BL/6 × 129SvEv | M:82 | 82 | 2.08 | NR | Min et al. (2008) |
| F:84 | |||||||
| −/− | M:58 | 128 | 0.89 | NR | |||
| F:45 | |||||||
| Trp63 | WT | K5CrePR1 | NR | 74 | 2.23 | 3.00 | Keyes et al. (2005) |
| +/− | NR | 104 | 1.74 | 2.25 | |||
| Xrcc5 | WT | NR | 47 | 2.04 | 2.40 | Vogel et al. (1999) | |
| Ku86 −/− | NR | 89 | 0.79 | 1.75 | |||
| Xrcc6 | WT | M & F | 27 | 2.17 | 2.80 | Li et al. (2007) | |
| Ku70 −/− | M & F | 43 | 0.69 | 1.45 |
t50, median life span; tmax, maximum life span; yr, year; WT, wild-type; M, male; F, female; Tg, transgenic; NR, not reported;
Mortality data were extracted from published studies. WebPlotDigitizer, an online graph digitizer application (https://automeris.io/WebPlotDigitizer/), was used to extract raw data from survival graphs in some cases. The digital imaging software package PaintShop Pro X3 (Corel Corporation, Ottawa, Canada) was used to extract age-specific survival data from published survivorship graphs (usually in the form of Kaplan–Meier survivorship curves). Percent survival was extracted at regular time intervals over the linear phase of these plots. Interval length was selected for each study individually to maximize the number of consecutive time points analyzed within this exponential phase.
Demographic analysis
The same methods and computer programs for the data gathering and analysis of de Magalhães et al. (2005) were used. Once the mortality data were collected, the age-specific mortality (qx) was calculated as the number of mice alive (T1) at the beginning of a given time interval minus the number of mice alive (T2) at the end of that same time interval divided by T1 (i.e., (T1 − T2)/T1 = qx). The hazard rate (hz) for each individual time interval was calculated as hz = (T1 − T2)/((T1 + T2)/2) or the number of animals dying in the interval divided by the average number of individuals alive in the interval. The aging rate was then calculated through use of the Gompertz equation: Rm(t) = R0eαt; where Rm(t) is the chance of dying (the hazard rate) at age t, R0 is the nonexponential factor in mortality, and α is the exponential parameter (Finch et al. 1990). The Gompertz model was used because, as de Magalhães et al. (2005) asserted, the sample size (number of mice) in the majority of these experiments was small, meaning that other logarithmic methods of fitting models to mortality data may not provide the accuracy that the Gompertz model gives while retaining its simplicity. This point held true for the majority of genetic interventions tested herein and so it was decided that use of the Gompertz model remained a viable option for this analysis. From the Gompertz model, the weighted regression line (weighted by the number of animals dying at each interval) was calculated by ln(Rm(t)) = ln(R0) + αt, which will also give the MRDT as 0.693/α. To compare α between a given genetic intervention cohort and wild-type (WT) mice from the same lineage, a “dummy variables” test was employed as described (de Magalhães et al. 2005). As in de Magalhães et al. (2005), the aim of this research was not to find the best fit model to describe the whole of the mortality curves, but rather to find if any previously published genetic interventions have a statistically significant effect on the exponential increase in mortality (α) and hence on the demographic rate of aging. Consequently, the simpler, nested Gompertz model was preferred (de Magalhães et al. 2005). Besides, data were only analyzed from the onset of the exponential increase in mortality, though typically < 10% of animals were left out.
Statistical analysis
A “dummy variables” method was used to compare the slopes of the hazard functions obtained through linear regression for the WT and test cohorts and a two-tailed Student’s t-test was applied to evaluate whether they were significantly different (P < 0.05). Analysis was performed in SPSS version 22 (IBM) using our previous scripts (de Magalhães et al. 2005) (code available in the supplemental material and at http://genomics.senescence.info/software/demographic.html).
Results
The Gompertz law of mortality describes the exponential increase in mortality rate with age. Modified versions of this function exist to model mortality deceleration observed at very young and very old ages; however, the simplest form was chosen to fit the mouse survivorship data in this study because the mouse cohorts are often small (n < 50). The basic Gompertz model is described by only two parameters, so is more suitable for fitting data from smaller sample sizes (Pletcher 2002) and increases the ease of comparing aging rates between cohorts. Since mouse longevity data are largely presented in publications as Kaplan–Meier survival curves, the life spans were divided into discrete time intervals and then linear regression was used to calculate age-specific mortality rates for each interval (see Methods). This allowed an estimation of Gompertz parameters for each cohort that could be directly compared.
In this study, 54 previously published genetic manipulations that have been associated with alterations in mouse life span were analyzed; 30 manipulations previously reported as having a life span-extending effect (Table 1) and 24 that were previously reported as having a life span-reducing effect (Table 2). The aim of this study was to reassess genes that have been reported to regulate longevity in mice to ascertain which of them might exert this effect through regulating the rate of aging.
Analysis of life-extending gene manipulations
Of the 30 genetic manipulations previously reported as having life span-increasing effects, we found 13 genes to have a statistically significant effect on the demographic rate of aging (Table 3): BubR1, Cisd2, Dgat1, Fgf21, Ghrh, Gpx4, hMTH1, Irs2, mTOR, Sirt1, Slc13a1, Surf1, and Tert. However, surprisingly only two of these genes (Cisd2 and hMTH1) retarded the demographic rate of aging. Full survival data are provided in the supplemental material (Table S1 in File S1).
Table 3
| Gene or protein | Type | Strain | α | α SE | ln(R0) | r2 | MRDT (yr) |
|---|---|---|---|---|---|---|---|
| Adcy5 | WT | 129/SvJ-C57BL/6 | 3.87 | 0.94 | −9.61 | 0.71 | 0.18 |
| −/− | 5.95 | 0.34 | −17.20 | 0.97 | 0.12 | ||
| Agrt1a | WT | C57BL/6 × 129/SvEv | 5.20 | 1.21 | −11.15 | 0.86 | 0.13 |
| −/− | 3.95 | 1.30 | −10.81 | 0.70 | 0.18 | ||
| Arf/p53 | WT | C57BL/6J | 2.88 | 0.32 | −7.59 | 0.94 | 0.24 |
| s-Arf/p53 | 2.80 | 0.44 | −8.05 | 0.86 | 0.25 | ||
| Atg5 | WT | C57BL/6 | 7.30 | 5.48 | −14.40 | 0.47 | 0.10 |
| Atg5-Tg | 8.92 | 1.31 | −21.54 | 0.82 | 0.08 | ||
| BubR1a | WT | C57BL/6-SV129 | 1.62 | 0.16 | −5.42 | 0.92 | 0.43 |
| mBubR1-Tg | 2.61 | 0.097 | −7.48 | 0.93 | 0.27 | ||
| Cat | WT | 4033 | 2.30 | 0.18 | −6.40 | 0.81 | 0.30 |
| MCAT | C57BL/6J | 3.00 | 0.59 | −8.80 | 0.68 | 0.23 | |
| WT | 4403 | 2.00 | 0.21 | −5.93 | 0.62 | 0.35 | |
| MCAT | 2.20 | 0.20 | −7.41 | 0.78 | 0.32 | ||
| Cisd2a | WT male | C57BL/6 | 7.67 | 2.86 | −17.79 | 0.71 | 0.09 |
| Cisd2-Tg male | 6.96 | 1.84 | −19.39 | 0.83 | 0.10 | ||
| WT female | 6.62 | 2.27 | −14.43 | 0.90 | 0.10 | ||
| Cisd2-Tg female | 2.40 | 0.59 | −7.10 | 0.74 | 0.29 | ||
| Clk-1 | WT | 129Sv/J | 3.09 | 0.64 | −6.05 | 0.89 | 0.22 |
| +/− | 2.49 | 0.92 | −5.95 | 0.64 | 0.28 | ||
| Dgat1a | WT | C57BL/6J | 1.92 | 0.53 | −5.12 | 0.66 | 0.36 |
| −/− | 3.56 | 0.20 | −9.63 | 0.99 | 0.19 | ||
| Esp8 | WT | C57BL/6 | 1.50 | 0.22 | −3.70 | 0.73 | 0.46 |
| −/− | 1.98 | 0.18 | −5.15 | 0.81 | 0.35 | ||
| FGF21a | WT | C57Bl/6J | 1.82 | 0.35 | −5.66 | 0.82 | 0.38 |
| FGF21-Tg | 2.04 | 0.40 | −7.89 | 0.87 | 0.34 | ||
| Ghrha | WT | C57BL/6 × 129SV | 1.31 | 0.18 | −3.55 | 0.90 | 0.53 |
| KO | 1.53 | 0.17 | −5.12 | 0.94 | 0.45 | ||
| Gpx4a | WT | C57BL/6 | 3.11 | 1.08 | −9.18 | 0.73 | 0.22 |
| +/– | 5.43 | 0.72 | −15.77 | 0.95 | 0.13 | ||
| hMTH1a | WT | C57BL/6 | 3.50 | 0.65 | −8.78 | 0.81 | 0.20 |
| hMTH1-Tg | 2.29 | 0.47 | −6.91 | 0.72 | 0.30 | ||
| Htt | WT | 129/Sv- | 2.30 | 1.01 | −6.03 | 0.56 | 0.30 |
| ΔQ /ΔQ | 1.80 | 0.51 | −5.19 | 0.81 | 0.39 | ||
| Igf1 | WT | FVB | 1.06 | 0.26 | −3.02 | 0.74 | 0.65 |
| Igf1 | 1.35 | 0.25 | −4.43 | 0.77 | 0.51 | ||
| IκB-α | MBH-GFP | C57BL/6 | 4.15 | 0.74 | −10.75 | 0.65 | 0.17 |
| MBH-DN IB-α | 4.43 | 0.76 | −12.86 | 0.62 | 0.16 | ||
| Irs1 | WT (female) | C57BL/6 | 2.18 | 0.26 | −5.00 | 0.80 | 0.32 |
| −/− (female) | 1.94 | 0.31 | −6.45 | 0.71 | 0.36 | ||
| Irs2a | WT | C57BL/6J | 2.99 | 0.13 | −7.29 | 0.87 | 0.23 |
| Brain-specific +/− | 6.37 | 0.33 | −17.73 | 0.85 | 0.11 | ||
| Mif | WT | C57BL/6J × 129/SvJ | 2.45 | 0.40 | −6.08 | 0.84 | 0.28 |
| −/− | 1.90 | 0.31 | −6.02 | 0.82 | 0.37 | ||
| mTORa | WT | 129 × C57BL/6 | 1.47 | 0.45 | −4.46 | 0.68 | 0.47 |
| KO | 2.41 | 0.15 | −6.94 | 0.98 | 0.29 | ||
| Myc | WT female | C57BL/6 | 2.19 | 0.55 | −5.88 | 0.76 | 0.32 |
| +/– female | 3.19 | 1.05 | −9.64 | 0.70 | 0.22 | ||
| WT male | 2.53 | 0.21 | −7.06 | 0.96 | 0.27 | ||
| +/– male | 2.55 | 0.36 | −7.75 | 0.90 | 0.27 | ||
| PAPP-A | WT | C57BL6 × 129SV/E | 2.33 | 0.35 | −5.28 | 0.71 | 0.30 |
| −/− | 2.67 | 0.32 | −7.39 | 0.80 | 0.26 | ||
| Pten | WT | C57BL/6 × CBA | 2.33 | 0.14 | −5.83 | 0.97 | 0.30 |
| Pten-Tg | 2.52 | 0.24 | −7.07 | 0.95 | 0.27 | ||
| RpS6K1 | WT | C57BL/6 | 1.81 | 0.069 | −4.64 | 0.93 | 0.38 |
| −/− | 1.73 | 0.056 | −5.30 | 0.91 | 0.40 | ||
| RIIβ | WT | C57BL/6 (males) | 3.00 | 0.31 | −7.68 | 0.85 | 0.23 |
| RIIB −/− | 3.55 | 0.34 | −10.09 | 0.88 | 0.20 | ||
| Sirt1a | WT | C57BL/6 | 2.45 | 0.49 | −7.20 | 0.81 | 0.28 |
| Brain-specific Tg | 3.63 | 0.63 | −10.71 | 0.83 | 0.19 | ||
| Slc13a1a | WT (male) | C57BL/6J × 129/SV | 1.79 | 0.37 | −4.34 | 0.83 | 0.39 |
| Nas1 −/− (male) | 1.70 | 0.47 | −5.02 | 0.72 | 0.41 | ||
| WT (female) | 0.87 | 0.13 | −2.95 | 0.86 | 0.80 | ||
| Nas1 −/− (female) | 1.69 | 0.29 | −4.31 | 0.85 | 0.41 | ||
| Surf1a | WT | BDF1 × cre | 2.19 | 0.39 | −4.81 | 0.86 | 0.32 |
| −/− | 3.41 | 0.47 | −7.82 | 0.96 | 0.20 | ||
| TgTerta | Sp53 | C57BL/6 × DBA/2 | 2.74 | 0.33 | −6.51 | 0.95 | 0.25 |
| Sp53a /TgTert | 3.34 | 0.41 | −8.14 | 0.94 | 0.21 | ||
| Sp53/Sp16/SArf | 1.47 | 0.58 | −4.86 | 0.52 | 0.47 | ||
| Sp53/Sp16/SArf/TgTert | 2.08 | 0.65 | −7.24 | 0.77 | 0.33 |
α, age-specific exponential coefficient; ln(R0), nonexponential factor in mortality; MRDT, mortality rate doubling time as in 0.693/α; yr, year; WT, wild-type; Tg, transgenic; KO, knockout.
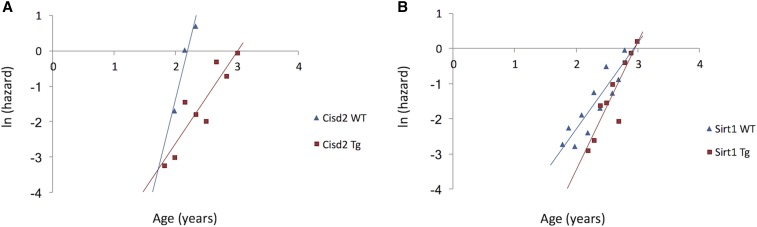
A few notable examples are worth emphasizing. Transgenic expression of Cisd2 in female mice produced persistent expression of the Cisd2 protein in contrast to levels in WT mice, which diminished with age (Wu et al. 2012). Our analysis showed that this resulted in a nearly twofold higher MRDT compared to that of female WT controls, consistent with a difference between the Gompertz curve gradients (Figure 1A). This suggests that Cisd2 regulates the demographic rate of aging. However, the nature of the role of Cisd2 in aging is confused by the fact that reduced expression of Cisd2 in heterozygous and double-knockout female mice from the same study also resulted in a significantly increased MRDT (see Table 4), suggesting that reduced Cisd2 expression also slowed the demographic rate of aging.
Natural logarithm of mortality rates for wild-type (WT, blue diamonds) and experimental transgenic (Tg, red squares) cohorts. Lines represent estimated adult mortality trajectories based on Gompertz parameters for the WT (blue lines) and experimental (red lines) cohorts. (A) Cisd2. (B) Sirt1.
Table 4
| Gene or protein | Type | Strain | α | α SE | ln(R0) | r2 | MRDT (yr) |
|---|---|---|---|---|---|---|---|
| Aag, Atm, Mgmta | WT | C57BL/6 | 3.81 | 0.82 | −8.83 | 0.78 | 0.18 |
| Aag –/–a | 2.20 | 1.04 | −5.38 | 0.53 | 0.31 | ||
| Mgmt –/– | 3.08 | 0.50 | −7.65 | 0.86 | 0.23 | ||
| Atm –/–a | 2.21 | 1.03 | −2.39 | 0.48 | 0.31 | ||
| Aag –/– Mgmt –/–a | 1.66 | 0.41 | −4.38 | 0.65 | 0.42 | ||
| ATR | WT | N/A | N/A | N/A | N/A | N/A | N/A |
| ATRs/s | 6.46 | 1.33 | −3.38 | 0.86 | 0.11 | ||
| Brca1 | WT | 129O1a × C57BL/6J | 4.59 | 0.36 | −10.01 | 0.98 | 0.15 |
| +/− (female) | 3.96 | 0.48 | −8.06 | 0.97 | 0.17 | ||
| Bub3 + Rae1 | WT | 129Sv/E × C57BL/6 | 1.54 | 0.34 | −4.86 | 0.91 | 0.45 |
| +/−, +/− | 1.16 | 0.49 | −3.72 | 0.61 | 0.60 | ||
| Casp2a | WT | C57BL/6 | 1.46 | 0.31 | −5.39 | 0.79 | 0.47 |
| −/− | 3.20 | 0.44 | −9.44 | 0.91 | 0.22 | ||
| Cd42 GAPa | WT | C57BL/6+/− 129/Sv | 3.39 | 1.11 | −8.16 | 0.61 | 0.20 |
| −/− | 1.24 | 1.44 | −2.89 | 0.87 | 0.56 | ||
| Cisd2a | WT | C57BL/6 (B6) | 3.42 | 0.81 | −8.37 | 0.64 | 0.20 |
| Cisd2−/− | 1.02 | 0.36 | −2.10 | 0.66 | 0.68 | ||
| Cisd2a | WT male | C57BL/6 | 7.67 | 2.86 | −17.79 | 0.71 | 0.09 |
| +/– malea | 4.06 | 1.26 | −9.68 | 0.68 | 0.17 | ||
| –/– male | 5.56 | 1.11 | −11.06 | 0.86 | 0.12 | ||
| WT female | 6.62 | 2.27 | −14.43 | 0.90 | 0.10 | ||
| +/– femalea | 2.50 | 0.49 | −5.69 | 0.81 | 0.28 | ||
| –/– femalea | 2.44 | 0.40 | −5.16 | 0.84 | 0.28 | ||
| DNA pol β | WT | C57BL/6 | 3.06 | 0.59 | −9.42 | 0.79 | 0.23 |
| +/– | 3.11 | 0.65 | −9.11 | 0.82 | 0.22 | ||
| Fgf-23 | −/− | Sv129J | 6.48 | N/A | −1.76 | 0.41 | 0.11 |
| Fn1a | WT | C57BL/6 | 1.49 | 0.48 | −5.22 | 0.62 | 0.46 |
| Fn1EDA −/− | 2.73 | 0.56 | −6.01 | 0.82 | 0.25 | ||
| Htr1ba | WT | 4.95 | 0.83 | −13.74 | 0.90 | 0.14 | |
| −/− | 4.06 | 0.46 | −9.56 | 0.94 | 0.17 | ||
| HtrA2/Omi | mnd2/mnd2;Tg | C57BL/6J | N/A | N/A | N/A | N/A | N/A |
| mnd2/+;Tg | 5.38 | 1.69 | −7.16 | 0.84 | 0.13 | ||
| IKK-βa | MBH-GFP | C57BL/6 | 4.78 | 1.09 | −12.43 | 0.76 | 0.15 |
| N/Ikbkbl/l | 6.03 | 0.84 | −14.37 | 0.90 | 0.12 | ||
| junDa | WT | N/A | 2.67 | 0.77 | −5.85 | 0.75 | 0.26 |
| JunD−/− | N/A | 3.89 | 0.57 | −7.47 | 0.90 | 0.18 | |
| Msh2 | WT | 129S/SvEvTac | N/A | N/A | N/A | N/A | N/A |
| −/− | 1.69 | N/A | −2.69 | 0.26 | 0.41 | ||
| Pasg | −/− | 129/SvJ/C57BL/6J | 64.15 | N/A | −2.55 | 0.75 | 0.01 |
| Pparga | WT | 4.43 | 0.53 | −11.02 | 0.93 | 0.16 | |
| Pparg2−/− | 2.59 | 0.29 | −6.93 | 0.92 | 0.27 | ||
| Sirt7 | WT | C57Bl/6 × 129Sv | N/A | N/A | N/A | N/A | N/A |
| −/− | 2.16 | −2.01 | 0.71 | 0.32 | |||
| Socs2a | WT | C57BL/6J × FVB | 2.14 | 0.40 | −5.16 | 0.85 | 0.32 |
| hg/hg | 1.43 | 0.33 | −2.69 | 0.73 | 0.48 | ||
| Stub1a | WT | C57BL/6 × 129SvEv | 0.80 | 0.40 | −3.78 | 0.44 | 0.87 |
| −/− | 1.31 | 0.44 | −2.46 | 0.59 | 0.53 | ||
| Trp63a | WT | K5CrePR1 | 2.90 | N/A | −7.47 | 0.86 | 0.24 |
| +/− | 1.75 | N/A | −3.72 | 0.83 | 0.40 | ||
| Xrcc5a | WT | 2.68 | 0.35 | −5.84 | 0.94 | 0.26 | |
| Ku86 −/− | 1.63 | 0.38 | −2.34 | 0.70 | 0.43 | ||
| Xrcc6a | WT | 1.92 | 0.30 | −4.98 | 0.89 | 0.36 | |
| Ku70 −/− | 1.39 | 0.44 | −1.49 | 0.83 | 0.50 |
α, age-specific exponential coefficient; ln(R0), nonexponential factor in mortality; MRDT, mortality rate doubling time as in 0.693/α; yr, year; WT, wild-type; N/A, not applicable; Tg, transgenic.
Most genes examined did not impact on MRDT, and a few even reduced the MRDT. Of note, regarding Sirt1, a 33% reduction in MRDT was observed for transgenic mice with brain-specific overexpression of this gene. This is supported by a visible change in the slope of the Gompertz curve (Figure 1B). It therefore appears that the increases in median and maximum life span reported (Satoh et al. 2013) are not mediated by a decrease in the rate of aging. Instead, it appears that the longevity extension in this cohort occurred through delayed onset of the exponential increase in mortality rate. Likewise, mice constitutively overexpressing both the tumor suppressor p53 and telomerase reverse transcriptase (Tert) exhibited enhanced 3-year survival rates compared to super-p53 mice expressing only the additional transgenic copy of p53 (Tomás-Loba et al. 2008). Interestingly, our analysis indicates that the MRDT of the former cohort was modestly reduced by 18% compared to that of the control mice, which suggests that the life-extending effect of Tert overexpression is due to a reduced R0 rather than to a slower demographic rate of aging. By contrast, mice overexpressing Tert in addition to p16, Arf, and p53 have a higher MRDT, although this difference was not statistically significant (Table 3).
Analysis of genes reported to reduce life span and/or accelerate aging
Interventions that reduce life span by increasing the rate of aging would be expected to reduce the MRDT. Of the 24 genes previously reported as having life span-reducing effects, we found 15 to have a statistically significant effect on the demographic rate of aging (Table 4): Aag/Atm/Mgmt, Casp2, Cisd2 (2× studies), Cdc42GAP, Fn1, Htr1b, IKK-β, JunD, Pparg, Socs2, Stub1, Trp63, Xrcc5, and Xrcc6. Five of these (Casp2, Fn1, IKK-β, JunD, and Stub1) accelerated demographic aging. Full survival data are provided in the supplemental material (Table S2 in File S1).
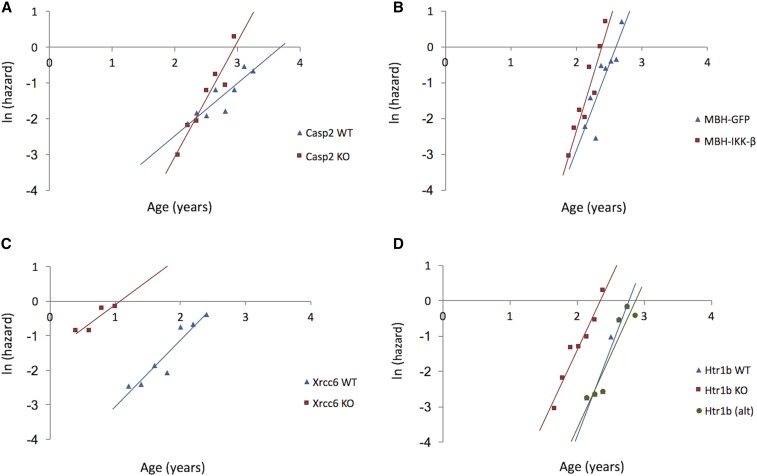
As before, a few notable examples are worth highlighting. Mice carrying a double knockout of Casp2 exhibited a 54% lower MRDT than WT littermates (Figure 2A). Therefore, as initially reported by Zhang et al. (2007), Casp2 accelerates the rate of aging in mice. Zhang et al. (2007) used several methods including a thorough detailing of the progression of age-related ailments (such as gradual hair and increased bone loss) to detail the rate of aging of the Casp2 knockout and their WT littermates, and our new research (using statistical demographic methods) has drawn the same conclusions.
Natural logarithm of mortality rates for wild-type (WT, blue diamonds) and experimental (red squares) cohorts. Lines represent estimated adult mortality trajectories based on Gompertz parameters for the WT (blue lines) and experimental (red lines) cohorts. KO, knockout. (A) Casp2; (B) MBH-IKK-β; (C) Xrcc6; and (D) Htr1b with the green line and circles representing an alternative for the WT cohort (see text for details).
Another gene manipulation that significantly reduced MRDT involved IκB kinase-β (IKK-β), which is involved in the activation of NF-κB. NF-κB activity in the hypothalamus of mice increases with age and mice expressing constitutively active IKK-β in the mediobasal hypothalamus (MBH) exhibited shortened life spans (Zhang et al. 2013). The MRDT for MBH-IKK-β mice was reduced by 21% compared to that of control mice. The fitted Gompertz curves showed an increased gradient (Figure 2B), providing evidence that activated hypothalamic NF-κB promotes faster aging. As the authors note, this is interesting because it suggests that a single organ, the hypothalamus, is important in regulating aging of the whole animal.
As in life-extending interventions, most genes did not impact on MRDT and some even had an opposite effect than expected. For example, Xrcc6 −/− (also known as Ku70 −/−) mice had shortened life spans compared to WT controls (Li et al. 2007). Fitting the survival data from this study produced mortality curves with visibly different starting ages (Figure 2C) and the MRDT of the Xrcc6 −/− mice was 38% larger than that of the WT cohort. This points to a slower demographic rate of aging in the Xrcc6 −/− mice. It should be noted that animals that died in the first 3 weeks were censored in the original study because Ku mutant mice frequently do not survive to weaning age. Including these animals would have altered the survival plots and might therefore have impacted on the outcome of this analysis.
Finally, reassessment of the survival data of Htr1b−/− mice showed that they have a 22% increased MRDT compared to WT controls. This can be seen as a slight difference in the slopes of the Gompertz curves (Figure 2D). Reduced longevity was reported in Htr1b−/− mice (Sibille et al. 2007) and our results support the conclusion that Htr1b deficiency produces a significantly decreased demographic rate of aging; however, the cohorts used in this study were small (n = 21 for WT and n = 24 for Htr1b−/−). If we increase the period from which data were extracted for the Htr1b−/− mice and exclude outliers of the Gompertz curve, the difference in MRDT is no longer significant. This result highlights the differences that one can obtain in this type of analysis by changing subjective parameters, in particular for smaller cohorts.
Longevity effects are driven by aging-independent mortality
Looking at our data set as a whole (Table 1, Table 2, Table 3, and Table 4), it is clear that studies are highly variable. Of note, cohort size ranges from 10 to 146 animals. Moreover, while the SD of median life span (t50) was only 13% for life span-extending manipulations (range 1.68–3.18 years), for life span-reducing manipulations it was 39% (0.01–2.62 years range). For maximum life span (tmax), SD was 11% for life span-extending manipulations (range 2.14–3.61 years) and 33% (0.07–3.51 years range) for life span-reducing manipulations. This is not surprising given that life-shortening manipulations can have greater effect sizes than life-extending manipulations, but it also introduces noise in demographic aging estimates, in particular for life span-shortening manipulations.
Even looking only at WT controls from the C57BL/6 strain (15 studies), the most common strain in our analysis, the range of t50 was 1.74–2.63 years while tmax ranged from 2.14 to 3.56. While tmax is influenced by cohort size, t50 is not and, therefore, this substantial variation for WT mice of the same genetic background suggests that considerable variation is introduced by differences in animal husbandry and stochastic factors. Relative SD for α and ln(R0) in C57BL/6 cohorts were, respectively, 57 and 42%.
We also investigated if, in life span-extending manipulations, there is a negative correlation between the t50 of the controls and the life extension effects (measured as the percentage t50 increase in the experimental cohort). Indeed, there is a moderate (r2 = 0.30) but statistically significant negative correlation (P-value = 0.002; n = 33), suggesting that effect sizes in longevity experiments could be influenced by the short life span of the controls.
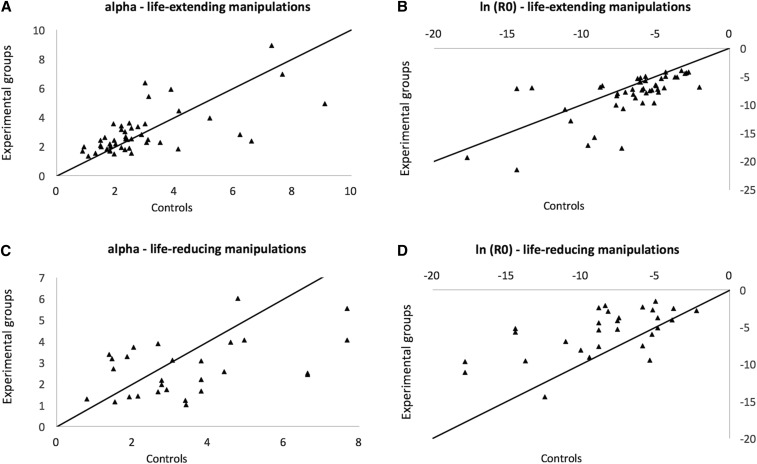
There was a strong negative correlation between α and ln(R0): r2 = 0.93 for life span-extending manipulations and r2 = 0.53 (after removing Pasg, which is an outlier; see Table 4) for life span-reducing manipulations. As such, increases in aging-independent mortality tend to be accompanied by a slower demographic aging rate, as observed before (de Magalhães et al. 2005).
Combining earlier results (de Magalhães et al. 2005) with the current analysis allows greater power to evaluate the usefulness of demographic analysis in aging. Therefore, we employed a data set with 63 manipulations of longevity: 41 life span-extending plus 22 life span-reducing manipulations (note that for this analysis we excluded manipulations for which we lacked demographic aging parameters for controls, which resulted in the exclusion of six life-reducing interventions from the previous analyses). Of note, we found that by and large an impact on longevity is caused by a change in the aging-independent mortality, which is observed both for life-extending (Figure 3, A and B) and life-reducing manipulations (Figure 3, C and D). And confirming the above-mentioned results, we observed that decreases in ln(R0) correlated with increases in α for life span-extending (r2 = 0.64) and life span-reducing manipulations (r2 = 0.64).
Discussion
The Gompertz function, used here to fit only the linear phase of the mortality trajectories, allowed for simple comparison between studies as it is described by just two parameters. Although it has been found that more complex adaptations of the Gompertz function [e.g., the logistic model (Pletcher et al. 2000)] provide a better fit for some whole-mortality curves (de Magalhães et al. 2005; Yen et al. 2008), the sample sizes usually reported for mouse life span experiments are insufficiently large to apply these models with sufficient accuracy.
Longevity is influenced by a number of factors, including age-independent mortality, age at the onset of senescence, and demographic rate of aging. In this study, we employed the Gompertz model to fit published mouse survival data and generate parameters that could be used to identify genes that influence the demographic rate of aging. Overall, only 7/54 genes were found to have a statistically significant effect on the demographic rate of aging as expected from longevity manipulations. These results suggest that only a relatively small proportion of interventions reported to affect longevity in mice do so through directly influencing the demographic rate of aging, in line with other, albeit smaller, studies (de Magalhães et al. 2005; Yen et al. 2008; Garratt et al. 2016; Hughes and Hekimi 2016). Surprisingly, 20/54 genes had a statistically significant impact on the demographic rate of aging in the opposite direction than would be expected for the published longevity effects. One possible explanation is that many mutations impacted on various parameters affecting longevity in nonlinear ways, and indeed we observed that increases in aging-independent mortality correlated with a slower demographic aging rate. For instance, Sirt1 deficiency extended life span but increased the demographic rate of aging; its effect appeared to be exerted instead by delaying the age of onset of mortality rate escalation. This highlights the complex relationship between life span and the demographic rate of aging. It is also possible that other confounding factors, like censored data or noise, in particular for smaller cohorts, influenced these results.
Another caveat of our approach concerns the number of mice used in some of the original studies, which ranged from 10 to 146 animals per cohort. While research reported here has attempted to compensate for this by using the Gompertz equation, which allows for small sample sizes, one cannot escape the low statistical power that accompanies such small sample sizes. Interestingly, caloric restriction has been shown to significantly retard the demographic rate of aging, but this was a large study with > 200 animals in total (de Magalhães et al. 2005). Therefore, caution must be taken when interpreting some of the results detailed here from studies with small sample sizes. Indeed, we observed that, in smaller experimental cohorts, subjective decisions in estimating Gompertz parameters can significantly affect the results, e.g., for Htr1b−/−.
Potential caveats of our analysis include the subjectivity of deciding the time at which analysis should begin and problems in the reporting of mouse survival data. As reviewed in detail by Ladiges et al. (2009), mouse longevity studies should adhere to certain standards to provide useful data. These include a sufficiently large cohort, high standards of pathogen-free animal husbandry to eliminate deaths from infectious disease, and separate reporting of male and female survival data. Unfortunately, life span data are often incompletely reported and, in many of the studies analyzed in this work, male and female data were not presented separately. Censoring of mice that died before a certain age in some studies may also have introduced noise into our results.
Our results provide insights regarding the role in aging of various genes. Of particular interest are the findings concerning Casp2 and Cisd2. We found that Casp2 deficiency increased the demographic rate of aging, which has not previously been proposed. Casp2 −/− mice had a similar median life span to WT and did not show elevated tumor incidence (Zhang et al. 2007). Besides, our results indicate that persistent expression of Cisd2 significantly reduced the demographic rate of aging. Further investigation has since strengthened the case for Cisd2's involvement in regulating the rate of aging by showing that it is involved in autophagy, mitochondrial function, and adipocyte differentiation (Wang et al. 2014), showing that it may influence several pathways thought to be important for aging.
Conclusions
Overall, we performed a demographic analysis of 54 mouse studies in which genetic manipulations significantly extended or reduced life span. We also combined our results with a previous report to perform an analysis of factors associated with longevity in mice. To our knowledge, this is the largest such study to date. Our main conclusions are: (1) most genetic manipulations of longevity in mice do so by modulating aging-independent mortality; (2) there is substantial variation in the life span of controls of the same strain across experiments; (3) studies in which the life span of the controls is short have a greater life span increase, emphasizing the importance of having adequate control groups; (4) mouse life span studies employing small cohorts can yield unreliable results; (5) life span-reducing experiments tend to be noisier and more difficult to analyze for demographic parameters than life-extending experiments; and (6) a greater aging-independent mortality is usually accompanied by a slower demographic aging rate.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.118.300821/-/DC1.
Acknowledgments
GenAge is funded by a Wellcome Trust grant (104978/Z/14/Z) to J.P.d.M. Current work in our laboratory is also supported by the Leverhulme Trust and the Methuselah Foundation. L.T. was funded by a Wellcome Trust 4-year PhD studentship.
Literature Cited
- Argmann C., Dobrin R., Heikkinen S., Auburtin A., Pouilly L., et al., 2009. Ppargamma2 is a key driver of longevity in the mouse. PLoS Genet. 5: e1000752 10.1371/journal.pgen.1000752 [PMC free article] [PubMed] [CrossRef] [Google Scholar]
- Baker D. J., Jeganathan K. B., Malureanu L., Perez-Terzic C., Terzic A., et al., 2006. Early aging-associated phenotypes in Bub3/Rae1 haploinsufficient mice. J. Cell Biol. 172: 529–540. 10.1083/jcb.200507081 [PMC free article] [PubMed] [CrossRef] [Google Scholar]
- Baker D. J., Dawlaty M. M., Wijshake T., Jeganathan K. B., Malureanu L., et al., 2013. Increased expression of BubR1 protects against aneuploidy and cancer and extends healthy lifespan. Nat. Cell Biol. 15: 96–102. 10.1038/ncb2643 [PMC free article] [PubMed] [CrossRef] [Google Scholar]
- Barardo D., Thornton D., Thoppil H., Walsh M., Sharifi S., et al., 2017. The DrugAge database of aging-related drugs. Aging Cell 16: 594–597. 10.1111/acel.12585 [PMC free article] [PubMed] [CrossRef] [Google Scholar]
- Benigni A., Corna D., Zoja C., Sonzogni A., Latini R., et al., 2009. Disruption of the Ang II type 1 receptor promotes longevity in mice. J. Clin. Invest. 119: 524–530. 10.1172/JCI36703 [PMC free article] [PubMed] [CrossRef] [Google Scholar]
- Cabelof D. C., Ikeno Y., Nyska A., Busuttil R. A., Anyangwe N., et al., 2006. Haploinsufficiency in DNA polymerase beta increases cancer risk with age and alters mortality rate. Cancer Res. 66: 7460–7465. 10.1158/0008-5472.CAN-06-1177 [PubMed] [CrossRef] [Google Scholar]
- Casellas J., Medrano J. F., 2008. Lack of Socs2 expression reduces lifespan in high-growth mice. Age (Dordr.) 30: 245–249. 10.1007/s11357-008-9064-1 [PMC free article] [PubMed] [CrossRef] [Google Scholar]
- Chen Y. F., Kao C. H., Chen Y. T., Wang C. H., Wu C. Y., et al., 2009. Cisd2 deficiency drives premature aging and causes mitochondria-mediated defects in mice. Genes Dev. 23: 1183–1194. 10.1101/gad.1779509 [PMC free article] [PubMed] [CrossRef] [Google Scholar]
- Conover C. A., Bale L. K., 2007. Loss of pregnancy-associated plasma protein A extends lifespan in mice. Aging Cell 6: 727–729. 10.1111/j.1474-9726.2007.00328.x [PubMed] [CrossRef] [Google Scholar]
- Dell’agnello C., Leo S., Agostino A., Szabadkai G., Tiveron C., et al., 2007. Increased longevity and refractoriness to Ca(2+)-dependent neurodegeneration in Surf1 knockout mice. Hum. Mol. Genet. 16: 431–444. 10.1093/hmg/ddl477 [PubMed] [CrossRef] [Google Scholar]
- De Luca G., Ventura I., Sanghez V., Russo M. T., Ajmone-Cat M. A., et al., 2013. Prolonged lifespan with enhanced exploratory behavior in mice overexpressing the oxidized nucleoside triphosphatase hMTH1. Aging Cell 12: 695–705. 10.1111/acel.12094 [PubMed] [CrossRef] [Google Scholar]
- de Magalhães J. P., 2005. Open-minded scepticism: inferring the causal mechanisms of human ageing from genetic perturbations. Ageing Res. Rev. 4: 1–22. 10.1016/j.arr.2004.05.003 [PubMed] [CrossRef] [Google Scholar]
- de Magalhães J. P., Faragher R. G., 2008. Cell divisions and mammalian aging: integrative biology insights from genes that regulate longevity. BioEssays 30: 567–578. 10.1002/bies.20760 [PubMed] [CrossRef] [Google Scholar]
- de Magalhães J. P., Cabral J. A., Magalhaes D., 2005. The influence of genes on the aging process of mice: a statistical assessment of the genetics of aging. Genetics 169: 265–274. 10.1534/genetics.104.032292 [PMC free article] [PubMed] [CrossRef] [Google Scholar]
- de Magalhães J. P., Wuttke D., Wood S. H., Plank M., Vora C., 2012. Genome-environment interactions that modulate aging: powerful targets for drug discovery. Pharmacol. Rev. 64: 88–101. 10.1124/pr.110.004499 [PMC free article] [PubMed] [CrossRef] [Google Scholar]
- Enns L. C., Morton J. F., Treuting P. R., Emond M. J., Wolf N. S., et al., 2009. Disruption of protein kinase A in mice enhances healthy aging. PLoS One 4: e5963 (erratum: PLoS One 5: DOI: 10.1371/annotation/c7cad2dc-1eca-487e-89ae-151a22d8a0b4). 10.1371/journal.pone.0005963 [PMC free article] [PubMed] [CrossRef] [Google Scholar]
- Fernandes M., Wan C., Tacutu R., Barardo D., Rajput A., et al., 2016. Systematic analysis of the gerontome reveals links between aging and age-related diseases. Hum. Mol. Genet. 25: 4804–4818. [PMC free article] [PubMed] [Google Scholar]
- Finch C. E., 1990. Longevity, Senescence, and the Genome. The University of Chicago Press, Chicago. [Google Scholar]
- Finch C. E., Pike M. C., 1996. Maximum life span predictions from the Gompertz mortality model. J. Gerontol. A Biol. Sci. Med. Sci. 51: B183–B194. 10.1093/gerona/51A.3.B183 [PubMed] [CrossRef] [Google Scholar]
- Finch C. E., Pike M. C., Witten M., 1990. Slow mortality rate accelerations during aging in some animals approximate that of humans. Science 249: 902–905. 10.1126/science.2392680 [PubMed] [CrossRef] [Google Scholar]
- Garratt M., Nakagawa S., Simons M. J., 2016. Comparative idiosyncrasies in life extension by reduced mTOR signalling and its distinctiveness from dietary restriction. Aging Cell 15: 737–743. 10.1111/acel.12489 [PMC free article] [PubMed] [CrossRef] [Google Scholar]
- Harper J. M., Wilkinson J. E., Miller R. A., 2010. Macrophage migration inhibitory factor-knockout mice are long lived and respond to caloric restriction. FASEB J. 24: 2436–2442. 10.1096/fj.09-152223 [PMC free article] [PubMed] [CrossRef] [Google Scholar]
- Hayflick L., 2000. The future of ageing. Nature 408: 267–269. 10.1038/35041709 [PubMed] [CrossRef] [Google Scholar]
- Hinkal G., Parikh N., Donehower L. A., 2009. Timed somatic deletion of p53 in mice reveals age-associated differences in tumor progression. PLoS One 4: e6654 10.1371/journal.pone.0006654 [PMC free article] [PubMed] [CrossRef] [Google Scholar]
- Hofmann J. W., Zhao X., De Cecco M., Peterson A. L., Pagliaroli L., et al., 2015. Reduced expression of MYC increases longevity and enhances healthspan. Cell 160: 477–488. 10.1016/j.cell.2014.12.016 [PMC free article] [PubMed] [CrossRef] [Google Scholar]
- Hughes B. G., Hekimi S., 2016. Different mechanisms of longevity in long-lived mouse and Caenorhabditis elegans mutants revealed by statistical analysis of mortality rates. Genetics 204: 905–920. 10.1534/genetics.116.192369 [PMC free article] [PubMed] [CrossRef] [Google Scholar]
- Jeng Y. M., Cai-Ng S., Li A., Furuta S., Chew H., et al., 2007. Brca1 heterozygous mice have shortened life span and are prone to ovarian tumorigenesis with haploinsufficiency upon ionizing irradiation. Oncogene 26: 6160–6166. 10.1038/sj.onc.1210451 [PubMed] [CrossRef] [Google Scholar]
- Kang S., Louboutin J. P., Datta P., Landel C. P., Martinez D., et al., 2013. Loss of HtrA2/Omi activity in non-neuronal tissues of adult mice causes premature aging. Cell Death Differ. 20: 259–269. 10.1038/cdd.2012.117 [PMC free article] [PubMed] [CrossRef] [Google Scholar]
- Kenyon C. J., 2010. The genetics of ageing. Nature 464: 504–512 (erratum: Nature 467: 622). 10.1038/nature08980 [PubMed] [CrossRef] [Google Scholar]
- Keyes W. M., Wu Y., Vogel H., Guo X., Lowe S. W., et al., 2005. p63 deficiency activates a program of cellular senescence and leads to accelerated aging. Genes Dev. 19: 1986–1999. 10.1101/gad.342305 [PMC free article] [PubMed] [CrossRef] [Google Scholar]
- Ladiges W., Van Remmen H., Strong R., Ikeno Y., Treuting P., et al., 2009. Lifespan extension in genetically modified mice. Aging Cell 8: 346–352. [PubMed] [Google Scholar]
- Lapointe J., Stepanyan Z., Bigras E., Hekimi S., 2009. Reversal of the mitochondrial phenotype and slow development of oxidative biomarkers of aging in long-lived Mclk1+/− mice. J. Biol. Chem. 284: 20364–20374. 10.1074/jbc.M109.006569 [PMC free article] [PubMed] [CrossRef] [Google Scholar]
- Laurent G., Solari F., Mateescu B., Karaca M., Castel J., et al., 2008. Oxidative stress contributes to aging by enhancing pancreatic angiogenesis and insulin signaling. Cell Metab. 7: 113–124. 10.1016/j.cmet.2007.12.010 [PubMed] [CrossRef] [Google Scholar]
- Li H., Vogel H., Holcomb V. B., Gu Y., Hasty P., 2007. Deletion of Ku70, Ku80, or both causes early aging without substantially increased cancer. Mol. Cell. Biol. 27: 8205–8214. 10.1128/MCB.00785-07 [PMC free article] [PubMed] [CrossRef] [Google Scholar]
- Li Q., Ren J., 2007. Influence of cardiac-specific overexpression of insulin-like growth factor 1 on lifespan and aging-associated changes in cardiac intracellular Ca2+ homeostasis, protein damage and apoptotic protein expression. Aging Cell 6: 799–806. 10.1111/j.1474-9726.2007.00343.x [PubMed] [CrossRef] [Google Scholar]
- Liu X., Jiang N., Hughes B., Bigras E., Shoubridge E., et al., 2005. Evolutionary conservation of the clk-1-dependent mechanism of longevity: loss of mclk1 increases cellular fitness and lifespan in mice. Genes Dev. 19: 2424–2434. 10.1101/gad.1352905 [PMC free article] [PubMed] [CrossRef] [Google Scholar]
- Markovich D., Ku M. C., Muslim D., 2011. Increased lifespan in hyposulfatemic NaS1 null mice. Exp. Gerontol. 46: 833–835. 10.1016/j.exger.2011.05.008 [PubMed] [CrossRef] [Google Scholar]
- Matheu A., Maraver A., Klatt P., Flores I., Garcia-Cao I., et al., 2007. Delayed ageing through damage protection by the Arf/p53 pathway. Nature 448: 375–379. 10.1038/nature05949 [PubMed] [CrossRef] [Google Scholar]
- Meira L. B., Calvo J. A., Shah D., Klapacz J., Moroski-Erkul C. A., et al., 2014. Repair of endogenous DNA base lesions modulate lifespan in mice. DNA Repair (Amst.) 21: 78–86. 10.1016/j.dnarep.2014.05.012 [PMC free article] [PubMed] [CrossRef] [Google Scholar]
- Min J. N., Whaley R. A., Sharpless N. E., Lockyer P., Portbury A. L., et al., 2008. CHIP deficiency decreases longevity, with accelerated aging phenotypes accompanied by altered protein quality control. Mol. Cell. Biol. 28: 4018–4025. 10.1128/MCB.00296-08 [PMC free article] [PubMed] [CrossRef] [Google Scholar]
- Murga M., Bunting S., Montana M. F., Soria R., Mulero F., et al., 2009. A mouse model of ATR-Seckel shows embryonic replicative stress and accelerated aging. Nat. Genet. 41: 891–898. 10.1038/ng.420 [PMC free article] [PubMed] [CrossRef] [Google Scholar]
- Muro A. F., Chauhan A. K., Gajovic S., Iaconcig A., Porro F., et al., 2003. Regulated splicing of the fibronectin EDA exon is essential for proper skin wound healing and normal lifespan. J. Cell Biol. 162: 149–160. 10.1083/jcb.200212079 [PMC free article] [PubMed] [CrossRef] [Google Scholar]
- Ortega-Molina A., Efeyan A., Lopez-Guadamillas E., Munoz-Martin M., Gomez-Lopez G., et al., 2012. Pten positively regulates brown adipose function, energy expenditure, and longevity. Cell Metab. 15: 382–394. 10.1016/j.cmet.2012.02.001 [PubMed] [CrossRef] [Google Scholar]
- Pletcher S. D., 2002. Mitigating the tithonus error: genetic analysis of mortality phenotypes. Sci. SAGE KE 2002: pe14. [PubMed] [Google Scholar]
- Pletcher S. D., Khazaeli A. A., Curtsinger J. W., 2000. Why do life spans differ? Partitioning mean longevity differences in terms of age-specific mortality parameters. J. Gerontol. A Biol. Sci. Med. Sci. 55: B381–B389. 10.1093/gerona/55.8.B381 [PubMed] [CrossRef] [Google Scholar]
- Pyo J. O., Yoo S. M., Ahn H. H., Nah J., Hong S. H., et al., 2013. Overexpression of Atg5 in mice activates autophagy and extends lifespan. Nat. Commun. 4: 2300 10.1038/ncomms3300 [PMC free article] [PubMed] [CrossRef] [Google Scholar]
- Ran Q., Liang H., Ikeno Y., Qi W., Prolla T. A., et al., 2007. Reduction in glutathione peroxidase 4 increases life span through increased sensitivity to apoptosis. J. Gerontol. A Biol. Sci. Med. Sci. 62: 932–942. 10.1093/gerona/62.9.932 [PubMed] [CrossRef] [Google Scholar]
- Razzaque M. S., Sitara D., Taguchi T., St-Arnaud R., Lanske B., 2006. Premature aging-like phenotype in fibroblast growth factor 23 null mice is a vitamin D-mediated process. FASEB J. 20: 720–722. 10.1096/fj.05-5432fje [PMC free article] [PubMed] [CrossRef] [Google Scholar]
- Satoh A., Brace C. S., Rensing N., Cliften P., Wozniak D. F., et al., 2013. Sirt1 extends life span and delays aging in mice through the regulation of Nk2 homeobox 1 in the DMH and LH. Cell Metab. 18: 416–430. 10.1016/j.cmet.2013.07.013 [PMC free article] [PubMed] [CrossRef] [Google Scholar]
- Schriner S. E., Linford N. J., Martin G. M., Treuting P., Ogburn C. E., et al., 2005. Extension of murine life span by overexpression of catalase targeted to mitochondria. Science 308: 1909–1911. 10.1126/science.1106653 [PubMed] [CrossRef] [Google Scholar]
- Selman C., Tullet J. M., Wieser D., Irvine E., Lingard S. J., et al., 2009. Ribosomal protein S6 kinase 1 signaling regulates mammalian life span. Science 326: 140–144. 10.1126/science.1177221 [PMC free article] [PubMed] [CrossRef] [Google Scholar]
- Sibille E., Su J., Leman S., Le Guisquet A. M., Ibarguen-Vargas Y., et al., 2007. Lack of serotonin1B receptor expression leads to age-related motor dysfunction, early onset of brain molecular aging and reduced longevity. Mol. Psychiatry 12: 1042–1056. 10.1038/sj.mp.4001990 [PMC free article] [PubMed] [CrossRef] [Google Scholar]
- Streeper R. S., Grueter C. A., Salomonis N., Cases S., Levin M. C., et al., 2012. Deficiency of the lipid synthesis enzyme, DGAT1, extends longevity in mice. Aging (Albany N.Y.) 4: 13–27. [PMC free article] [PubMed] [Google Scholar]
- Sun L. Q., Lee D. W., Zhang Q., Xiao W., Raabe E. H., et al., 2004. Growth retardation and premature aging phenotypes in mice with disruption of the SNF2-like gene, PASG. Genes Dev. 18: 1035–1046. 10.1101/gad.1176104 [PMC free article] [PubMed] [CrossRef] [Google Scholar]
- Sun L. Y., Spong A., Swindell W. R., Fang Y., Hill C., et al., 2013. Growth hormone-releasing hormone disruption extends lifespan and regulates response to caloric restriction in mice. Elife 2: e01098 10.7554/eLife.01098 [PMC free article] [PubMed] [CrossRef] [Google Scholar]
- Tacutu R., Craig T., Budovsky A., Wuttke D., Lehmann G., et al., 2013. Human ageing genomic resources: integrated databases and tools for the biology and genetics of ageing. Nucleic Acids Res. 41: D1027–D1033. 10.1093/nar/gks1155 [PMC free article] [PubMed] [CrossRef] [Google Scholar]
- Taguchi A., Wartschow L. M., White M. F., 2007. Brain IRS2 signaling coordinates life span and nutrient homeostasis. Science 317: 369–372. 10.1126/science.1142179 [PubMed] [CrossRef] [Google Scholar]
- Tocchetti A., Soppo C. B., Zani F., Bianchi F., Gagliani M. C., et al., 2010. Loss of the actin remodeler Eps8 causes intestinal defects and improved metabolic status in mice. PLoS One 5: e9468 10.1371/journal.pone.0009468 [PMC free article] [PubMed] [CrossRef] [Google Scholar]
- Tomás-Loba A., Flores I., Fernández-Marcos P. J., Cayuela M. L., Maraver A., et al., 2008. Telomerase reverse transcriptase delays aging in cancer-resistant mice. Cell 135: 609–622. 10.1016/j.cell.2008.09.034 [PubMed] [CrossRef] [Google Scholar]
- Vakhrusheva O., Smolka C., Gajawada P., Kostin S., Boettger T., et al., 2008. Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circ. Res. 102: 703–710. 10.1161/CIRCRESAHA.107.164558 [PubMed] [CrossRef] [Google Scholar]
- Vogel H., Lim D. S., Karsenty G., Finegold M., Hasty P., 1999. Deletion of Ku86 causes early onset of senescence in mice. Proc. Natl. Acad. Sci. USA 96: 10770–10775. 10.1073/pnas.96.19.10770 [PMC free article] [PubMed] [CrossRef] [Google Scholar]
- Wang C. H., Chen Y. F., Wu C. Y., Wu P. C., Huang Y. L., et al., 2014. Cisd2 modulates the differentiation and functioning of adipocytes by regulating intracellular Ca2+ homeostasis. Hum. Mol. Genet. 23: 4770–4785. 10.1093/hmg/ddu193 [PubMed] [CrossRef] [Google Scholar]
- Wang L., Yang L., Debidda M., Witte D., Zheng Y., 2007. Cdc42 GTPase-activating protein deficiency promotes genomic instability and premature aging-like phenotypes. Proc. Natl. Acad. Sci. USA 104: 1248–1253. 10.1073/pnas.0609149104 [PMC free article] [PubMed] [CrossRef] [Google Scholar]
- Wei K., Clark A. B., Wong E., Kane M. F., Mazur D. J., et al., 2003. Inactivation of Exonuclease 1 in mice results in DNA mismatch repair defects, increased cancer susceptibility, and male and female sterility. Genes Dev. 17: 603–614. 10.1101/gad.1060603 [PMC free article] [PubMed] [CrossRef] [Google Scholar]
- Wu C. Y., Chen Y. F., Wang C. H., Kao C. H., Zhuang H. W., et al., 2012. A persistent level of Cisd2 extends healthy lifespan and delays aging in mice. Hum. Mol. Genet. 21: 3956–3968. 10.1093/hmg/dds210 [PubMed] [CrossRef] [Google Scholar]
- Wu J. J., Liu J., Chen E. B., Wang J. J., Cao L., et al., 2013. Increased mammalian lifespan and a segmental and tissue-specific slowing of aging after genetic reduction of mTOR expression. Cell Rep. 4: 913–920. 10.1016/j.celrep.2013.07.030 [PMC free article] [PubMed] [CrossRef] [Google Scholar]
- Yan L., Vatner D. E., O’Connor J. P., Ivessa A., Ge H., et al., 2007. Type 5 adenylyl cyclase disruption increases longevity and protects against stress. Cell 130: 247–258. 10.1016/j.cell.2007.05.038 [PubMed] [CrossRef] [Google Scholar]
- Yen K., Steinsaltz D., Mobbs C. V., 2008. Validated analysis of mortality rates demonstrates distinct genetic mechanisms that influence lifespan. Exp. Gerontol. 43: 1044–1051. 10.1016/j.exger.2008.09.006 [PubMed] [CrossRef] [Google Scholar]
- Zhang G., Li J., Purkayastha S., Tang Y., Zhang H., et al., 2013. Hypothalamic programming of systemic ageing involving IKK-beta, NF-kappaB and GnRH. Nature 497: 211–216. 10.1038/nature12143 [PMC free article] [PubMed] [CrossRef] [Google Scholar]
- Zhang Y., Padalecki S. S., Chaudhuri A. R., De Waal E., Goins B. A., et al., 2007. Caspase-2 deficiency enhances aging-related traits in mice. Mech. Ageing Dev. 128: 213–221. 10.1016/j.mad.2006.11.030 [PMC free article] [PubMed] [CrossRef] [Google Scholar]
- Zhang Y., Xie Y., Berglund E. D., Coate K. C., He T. T., et al., 2012. The starvation hormone, fibroblast growth factor-21, extends lifespan in mice. Elife 1: e00065 10.7554/eLife.00065 [PMC free article] [PubMed] [CrossRef] [Google Scholar]
- Zheng S., Clabough E. B., Sarkar S., Futter M., Rubinsztein D. C., et al., 2010. Deletion of the huntingtin polyglutamine stretch enhances neuronal autophagy and longevity in mice. PLoS Genet. 6: e1000838 10.1371/journal.pgen.1000838 [PMC free article] [PubMed] [CrossRef] [Google Scholar]