- Journal List
- HHS Author Manuscripts
- PMC7325601

T cell-expressed microRNA-155 reduces lifespan in a mouse model of age-related chronic inflammation
H. Atakan Ekiz
†Department of Pathology, University of Utah School of Medicine, 15 N Medical Drive East RM5520, Salt Lake City, UT, 84112, USA.
‡Huntsman Cancer Institute, University of Utah, 2000 Circle of Hope, Salt Lake City, UT, 84112, USA.
Andrew G. Ramstead
†Department of Pathology, University of Utah School of Medicine, 15 N Medical Drive East RM5520, Salt Lake City, UT, 84112, USA.
‡Huntsman Cancer Institute, University of Utah, 2000 Circle of Hope, Salt Lake City, UT, 84112, USA.
Soh-Hyun Lee
†Department of Pathology, University of Utah School of Medicine, 15 N Medical Drive East RM5520, Salt Lake City, UT, 84112, USA.
‡Huntsman Cancer Institute, University of Utah, 2000 Circle of Hope, Salt Lake City, UT, 84112, USA.
Morgan C. Nelson
†Department of Pathology, University of Utah School of Medicine, 15 N Medical Drive East RM5520, Salt Lake City, UT, 84112, USA.
‡Huntsman Cancer Institute, University of Utah, 2000 Circle of Hope, Salt Lake City, UT, 84112, USA.
Kaylyn M. Bauer
†Department of Pathology, University of Utah School of Medicine, 15 N Medical Drive East RM5520, Salt Lake City, UT, 84112, USA.
‡Huntsman Cancer Institute, University of Utah, 2000 Circle of Hope, Salt Lake City, UT, 84112, USA.
Jared A. Wallace
†Department of Pathology, University of Utah School of Medicine, 15 N Medical Drive East RM5520, Salt Lake City, UT, 84112, USA.
‡Huntsman Cancer Institute, University of Utah, 2000 Circle of Hope, Salt Lake City, UT, 84112, USA.
Ruozhen Hu
†Department of Pathology, University of Utah School of Medicine, 15 N Medical Drive East RM5520, Salt Lake City, UT, 84112, USA.
‡Huntsman Cancer Institute, University of Utah, 2000 Circle of Hope, Salt Lake City, UT, 84112, USA.
June L. Round
†Department of Pathology, University of Utah School of Medicine, 15 N Medical Drive East RM5520, Salt Lake City, UT, 84112, USA.
‡Huntsman Cancer Institute, University of Utah, 2000 Circle of Hope, Salt Lake City, UT, 84112, USA.
Jared Rutter
‡Huntsman Cancer Institute, University of Utah, 2000 Circle of Hope, Salt Lake City, UT, 84112, USA.
¶Department of Biochemistry, University of Utah School of Medicine, 15 N Medical Drive East, RM2100, Salt Lake City, UT, 84112, USA.
||Howard Hughes Medical Institute, University of Utah School of Medicine, Salt Lake City, UT, USA
Micah J. Drummond
†Department of Pathology, University of Utah School of Medicine, 15 N Medical Drive East RM5520, Salt Lake City, UT, 84112, USA.
#Department of Physical Therapy and Athletic Training, University of Utah, Salt Lake City, UT, USA
**Department of Nutrition and Integrative Physiology, University of Utah, Salt Lake City, UT, USA
Dinesh S. Rao
††Department of Pathology and Laboratory Medicine, University of California Los Angeles, Box 951732, 11-544 Factor, Los Angeles, CA, 90095, USA
Ryan M. O’Connell
†Department of Pathology, University of Utah School of Medicine, 15 N Medical Drive East RM5520, Salt Lake City, UT, 84112, USA.
‡Huntsman Cancer Institute, University of Utah, 2000 Circle of Hope, Salt Lake City, UT, 84112, USA.
Author Contributions: Conceived and designed the experiments: HAE, AGR, SHL, RH, JAW, RMO. Performed the experiments: HAE, AGR, SHL, RH, MCN, JAW, KMB. Analyzed the data: HAE, AGR, RH, SHL, JAW, DSR. Contributed reagents/analysis/supervision: JLR, JR, DSR, MJD, RMO. Wrote the paper: HAE, AGR, RMO. All authors have significant input in the execution of the research project and the preparation of the manuscript.
Associated Data
Abstract
Aging-related chronic inflammation is a risk factor for many human disorders through incompletely understood mechanisms. Aged mice deficient in microRNA-146a succumb to life-shortening chronic inflammation. In this study, we report that miR-155 in T cells contributes to shortened lifespan of miR-146a−/− mice. Using single-cell RNA sequencing and flow cytometry we found that miR-155 promotes the activation of effector T cell populations, including Tfh cells, and increases germinal center B cells and autoantibodies in mice aged over 15 months. Mechanistically, aerobic glycolysis genes are elevated in T cells during aging, and upon deletion of miR-146a, in a T cell miR-155-dependent manner. Finally, skewing T cell metabolism towards aerobic glycolysis by deleting mitochondrial pyruvate carrier (MPC) recapitulates age-dependent T cell phenotypes observed in miR-146a−/− mice revealing the sufficiency of metabolic reprogramming to influence immune cell functions during aging. Altogether, these data indicate that T cell-specific miRNAs play pivotal roles in regulating lifespan through their influences on inflammaging.
Introduction
Chronic inflammation in the absence of infection is a commonly observed phenomenon during aging, and often involves upregulation of inflammatory cytokines, elevated auto-antibody titers, and impaired or altered hematopoiesis (1). This chronic, low-grade activation of the immune system upon aging, termed “inflammaging”, is implicated in the development of several age-related deleterious conditions including heart disease, neurodegeneration, autoimmunity, metabolic diseases and cancer (2). Studies have shown that the immune system undergoes significant changes and is skewed toward an increased production of myeloid cells which are thought to drive key inflammatory processes during aging (3, 4). Additionally, the critical involvement of T and B lymphocytes in aging-related immune dysfunction were recently appreciated (5–7). However, despite the clear correlations between chronic inflammation and disease onset or decreased lifespan, both the underlying mechanisms that drive inflammaging and approaches to treat or prevent this process remain largely unknown (8). A better understanding of these mechanisms will be critical for improving both the quality and duration of life in continually expanding elderly populations.
One way in which chronic inflammation and immune dysfunction can be manifested in the elderly is through the expansion of memory CD4+ T cells with a T follicular helper (Tfh) cell phenotype (9). Tfh cells are a subset of CD4+ T cells which provide help to germinal center (GC) B cells and promote antibody production (10). While this process is critical for defense against pathogens and the effectiveness of vaccines, improper expansion of Tfh cells can lead to the development of autoantibodies and autoimmunity, such as in the case of lupus (11). Studies have shown that the prevalence of Tfh cells increases during aging, along with the levels of autoantibodies (12–15). While the role of these autoantibodies in the aging process remains to be clearly elucidated, they may contribute to autoimmune diseases seen in aging populations (14, 16). Thus, an increased understanding of the mechanisms that control Tfh biology as well as other aberrant T cell responses is needed to mitigate the negative impacts of autoantibody production and other inflammatory mediators during the aging process.
MicroRNAs (miRNAs) are among the critical regulatory molecules that modulate lymphocyte biology, including Tfh and B cell functions, during aging. Two well-described miRNAs, miR-146a and miR-155, can serve as an immune rheostat by exerting opposing functions (17, 18). Both miR-146a and miR-155 are induced in multiple immune cell subsets including T cells, B cells, macrophages, and dendritic cells upon activation (18–22). miR-146a serves as a negative regulator of the immune response by suppressing the expression of several proteins involved in the Toll-like receptor (TLR) and T cell receptor (TCR) pathways, such as TNF receptor-associated factor 6 (TRAF6) and IL-1 receptor-associated kinase 1 (IRAK1) (19, 23). In contrast, miR-155 enhances the immune response by targeting negative regulators including SHIP1, SOCS1, and SOCS3 (24–26). miR-146a and miR-155 expression can be dysregulated in various inflammatory and autoimmune conditions. For instance, miR-146a was found to be expressed at lower levels in peripheral blood leukocytes of lupus patients compared to the healthy controls (27). Furthermore, mice lacking miR-146a expression experience multiple immune pathologies upon aging, such as myeloproliferation, splenomegaly, bone marrow failure, leukemia and autoimmunity (20, 28). Interestingly, some of these phenotypes can be reversed when miR-155 is deleted in miR-146a−/− mice (20). Supporting its immune-activating roles in miR-146a deficient mice, overexpression of miR-155 in wild type mice led to chronic inflammation characterized by myeloproliferation, splenomegaly and bone marrow failure (29, 30). Previously, we have shown that miR-155 expression in T cells was increased in middle-aged (~7 months-old) miR-146a-deficient mice compared to wild-type (WT) controls, and that T cell-specific miR-155 mediated the autoimmune phenotype during this intermediate stage of the murine lifespan (20). These findings suggest that the interplay between miR-146a and miR-155 regulates the immunopathologies associated with aging and might impact overall lifespan.
In this study, we examined survival and immunopathology in older (~15–16 months-old) miR-146a-deficient mice and assessed whether T cell-specific deletion of miR-155 in this setting is able to lengthen lifespan and reduce chronic inflammation in these animals. Our data reveal that the loss of miR-146a results in the expansion of Tfh cells, multi-organ autoimmunity and bone marrow failure in aged mice, correlating with dramatically reduced survival. Importantly, T cell-specific deletion of miR-155 in miR-146a-deficient animals suppressed the autoimmune phenotype and significantly extended their lifespan. However, bone marrow phenotypes that occurred in miR-146a−/− mice, including some aspects of myeloid skewing and bone marrow hypocellularity, were not substantially impacted by the loss of T cell miR-155 at this advanced age. Mechanistically, lack of miR-146a induced the expression of aerobic glycolysis genes and overactivation of both CD4+ and CD8+ T cells in the aging immune system. Enhanced glycolysis gene expression was largely reversed upon T cell-specific deletion of miR-155 suggesting that miR-146a and miR-155 regulate the metabolic state of T cells during the aging process. Importantly, skewing T cell metabolism to aerobic glycolysis through T cell-specific deletion of the mitochondrial pyruvate carrier 1 (Mpc1) gene resulted in the expansion of activated T cells, including Tfh cells, in aging mice similar to the miR-146a−/− condition. Taken together, our work substantially expands our understanding of the critical functions performed by miR-146a and miR-155 during age-mediated immune dysfunction, including their potential to influence lifespan.
Materials and Methods
Study Design
The overall objective of the controlled laboratory experiments herein was to characterize the roles of T cell-expressed miR-155 in the process of age-related chronic inflammation. Sample sizes in experiments involving transgenic animals were determined by power analyses and through consulting with the Biostatistics Core at the University of Utah School of Medicine. Datapoints were not removed from analyses unless they meet stringent criteria determined by statistical outlier tests. The primary endpoint was selected to be ~15 months in experiments measuring survival differences between groups based on prior experience. To address sex as an experimental variable, we conducted experiments using both male and female mice and observed similar results. Experiments were repeated multiple times unless otherwise indicated and representative results are shown.
Mice
Mice described in these studies are on the C57BL/6 genetic background and were housed in the specific-pathogen-free animal facility on a 12-hour light/dark cycle at the University of Utah. miR-146a−/−, miR-146a−/− CD4-Cre miR-155fl/fl, and MPC1fl/fl mice were described previously (31, 32). Founder mouse strains were purchased from Jackson Laboratories. Both female and male age-matched mice were used in all experiments. All animal protocols were approved by the Institutional Animal Care and Use Committee at the University of Utah.
Processing of mouse splenocytes and bone marrow
Spleens were removed from mice, homogenized using the frosted end of microscope slides, and filtered through a 40μM filter to obtain single cell suspensions. Red blood cells (RBC) were then lysed with RBC lysis buffer (Biolegend) and single cells were resuspended in culture media (RPMI media containing 10% FBS, penicillin, and streptomycin). To collect bone marrow cells, femurs were isolated and bone marrow was flushed using PBS. Red blood cells were then lysed with RBC lysis buffer and samples were resuspended in culture medium. Organ cellularity was assessed by enumerating isolated cells using a hemocytometer.
Single Cell Transcriptomics
Splenic immune landscape was analyzed using 10X Genomics single cell transcriptomics. Briefly, CD45+ splenic lymphocytes (FSC-low, SSC-low) were sorted via flow cytometry as described previously (33). Representative spleens from 3 (young WT males) or 6 (15-months-old WT, miR-146a−/−, DKO males) were pooled per group and equal numbers of cells were partitioned followed by single cell 3` gene expression library preparation as per manufacturer’s recommendations. A 125-cycle paired-end sequencing run was performed using a HiSeq SBS kit (v4). After sequencing, data were processed using the CellRanger pipeline (10X Genomics) which showed at least 5700 cells were sequenced with a minimum of 116.5 million reads per sample and the 15,968 mean reads per cell across 4 separate runs. Data are further analyzed using Seurat single cell genomics R package (34). Different sequencing runs were aggregated using Seurat’s built-in functions and 10 metagene dimensions to simultaneously analyze all 4 experimental groups. Single cell clusters identified by the Seurat algorithm were annotated using our previously published algorithm (33) (available online at https://aekiz.shinyapps.io/CIPR/), and by performing differential expression analyses. Visualizations were generated using R programming environment and ggpubr and fgsea R packages.
ELISA for dsDNA autoantibodies and total antibody levels
Serum autoantibodies specific for dsDNA were analyzed using a commercial ELISA kit (BioVendor) according to manufacturer’s protocol. Briefly, serial dilutions of serum were added to dsDNA-coated 96 well plates. After 2 hours of incubation plates were washed and anti-dsDNA autoantibodies were spectrophotometrically detected using an HRP-conjugated anti-mouse IgG secondary antibody. A similar experimental approach was employed to quantify the total IgG levels in which anti-mouse IgG capture antibodies were used for plate coating.
Western Blots
To measure circulating autoantibodies specific for tissue antigens, serum was collected by spinning down blood isolated via cardiac puncture at 10000 rpm for 10 min. Target tissues including brains, kidneys and livers were collected after whole body perfusion with PBS. Tissues were then homogenized with a Dounce homogenizer in RIPA buffer with Protease inhibitor (Pierce). Tissues were then spun down 12000 rpm for 10 minutes and supernatant containing tissue proteome was collected. 30 ug of protein was run on a 10% SDS-Page gel, then transferred to nitrocellulose paper. After blocking with milk, a 1:100 dilution of serum was used as primary antibody for western blots. Following incubation of the membranes with serum, a 1:2000 dilution of an HRP-conjugated anti-mouse IgG antibody (Jackson Immunoresearch) was used as the secondary antibody to reveal self-reactive antibodies upon treatment with ECL solution.
Flow cytometry
Fluorophore-conjugated antibodies were purchased from eBioscience (Thermo) or Biolegend and used to stain RBC-depleted splenocytes or bone marrow cells. The following clones were used: B220 (RA3–6B2), CD3e (142–2c11), CD4 (GK1.5), CD8a (53–6.7), CD11b (M1/70), CD44 (IM7), CD45 (30-F11), CD62L (MEL-14), CD69 (H1.2 F3), CXCR5 (L138 D7), Fas (Jo2), GL7 (GL7), IgD (11–26c.2a), ICOS (7E.17G9), PD-1 (RMP1–30), Ter119 (TER119). Cells were stained on ice in FACS buffer (PBS containing 2% FBS and 0.01% sodium azide). After staining, cells were washed two times with FACS buffer and were fixed for 15 mins in 1% formalin for future analysis. Stained cells were analyzed with a BD LSR Fortessa flow cytometer equipped with 4 lasers. Data were analyzed using FlowJo software. CD3e+CD8a+ or CD3e+CD4+ gates were used to define T cells in the analyses. Tfh cells were defined by positively gating on PD1 and CXCR5 or PD1 and ICOS within the CD3e+CD4+ T cell subset. GC B cell subset was defined as Fas+GL7+ of the B220+IgD(low) cells in the spleen. Flow cytometry gating was applied after excluding debris and doublets using the forward/side scatter data.
Statistics
Survival data were analyzed using log-rank test between pairs of groups. Organ cellularity and flow cytometry data were analyzed by ANOVA with Tukey’s correction using GraphPad Prism 7. In scRNAseq experiments, gene expression differences among groups were calculated using Wilcoxon test with a Benjamini-Hochberg correction for multiple testing. GSEA was performed using fgsea R package (35) and p-values were corrected for multiple testing using Benjamini-Hochberg method. In experiments involving two groups (such as Mpc1 knockout vs floxed control group) student’s t-test was used for statistical comparisons. The p values are represented by asterisks as follows: ns, not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Results
Loss of miR-146a results in increased mortality during aging which is rescued by T cell-specific deletion of miR-155
Previous work by our laboratory and others have demonstrated that miR-146a is responsible for dampening chronic inflammation during aging. Further, upon reaching middle-age (~7 months), mice lacking miR-146a begin to develop chronic inflammation that could be partially reversed by deleting miR-155 in T cells (20, 36–38). To determine if miR-155 function within T cells could impact the actual lifespan of miR-146a−/− mice, we examined both female and male mice from three groups for ~15 months: 1) miR-155 fl/fl (wild-type, WT, n=44), 2) miR-146a−/− (miR-146a KO, n=34), and 3) CD4-Cre miR-155 fl/fl miR-146a−/− (miR-146a-KO/155-T cell Conditional KnockOut (TCKO), or double knockout, DKO, n=55). As expected based on previous observations (23, 37, 39), miR-146a−/− mice exhibited a significantly reduced survival frequency compared to WT controls (Figure 1a–c). Surprisingly, deleting miR-155 from T cell subsets in these animals significantly extended their lifespan, albeit not back to WT levels (Figure 1a–c). These observations were consistent in both female and male mice although the effect was more pronounced in the female cohort (Figure 1a). Consistent with the systemic inflammation and myeloproliferative disease, the loss of miR-146a resulted in a lower body weight and increased spleen mass (Figure 1d, ,e).e). T-cell specific deletion of miR-155 restored the body weight back to WT levels and significantly reduced the splenomegaly observed in both male and female miR-146a−/− mice (Figure 1d, ,e).e). The same trends were observed when spleen cellularity was examined (Figure 1f). Loss of miR-146a was characterized by the dysregulation of hematopoiesis and a decline in the numbers of hematopoietic cells in the bone marrow (Figure 1g) (23). Interestingly, deletion of miR-155 in T cells did not affect the bone marrow hypocellularity, suggesting its survival benefit is not due to a better maintenance of the hematopoietic stem cells in the bone marrow. We further examined the cellular composition of the bone marrow and noted a significant reduction in the numbers of B220+ B cells (Figure 1h), Ter119+ erythroid cells (Figure 1i), and CD11b+ myeloid cells (Figure 1j) in aged miR-146a−/− mice compared to WT counterparts. Consistent with the bone marrow cellularity data, these phenotypes were not rescued in the DKO mice at this time point. These findings suggest that the rescue of the miR-146a−/− phenotype by the deletion of miR-155 in T cells is partial because survival is likely mediated through at least two independent miR-146a regulated pathways: 1) T cell-independent regulation of hematopoiesis and 2) T cell miR-155-dependent regulation of autoimmunity.
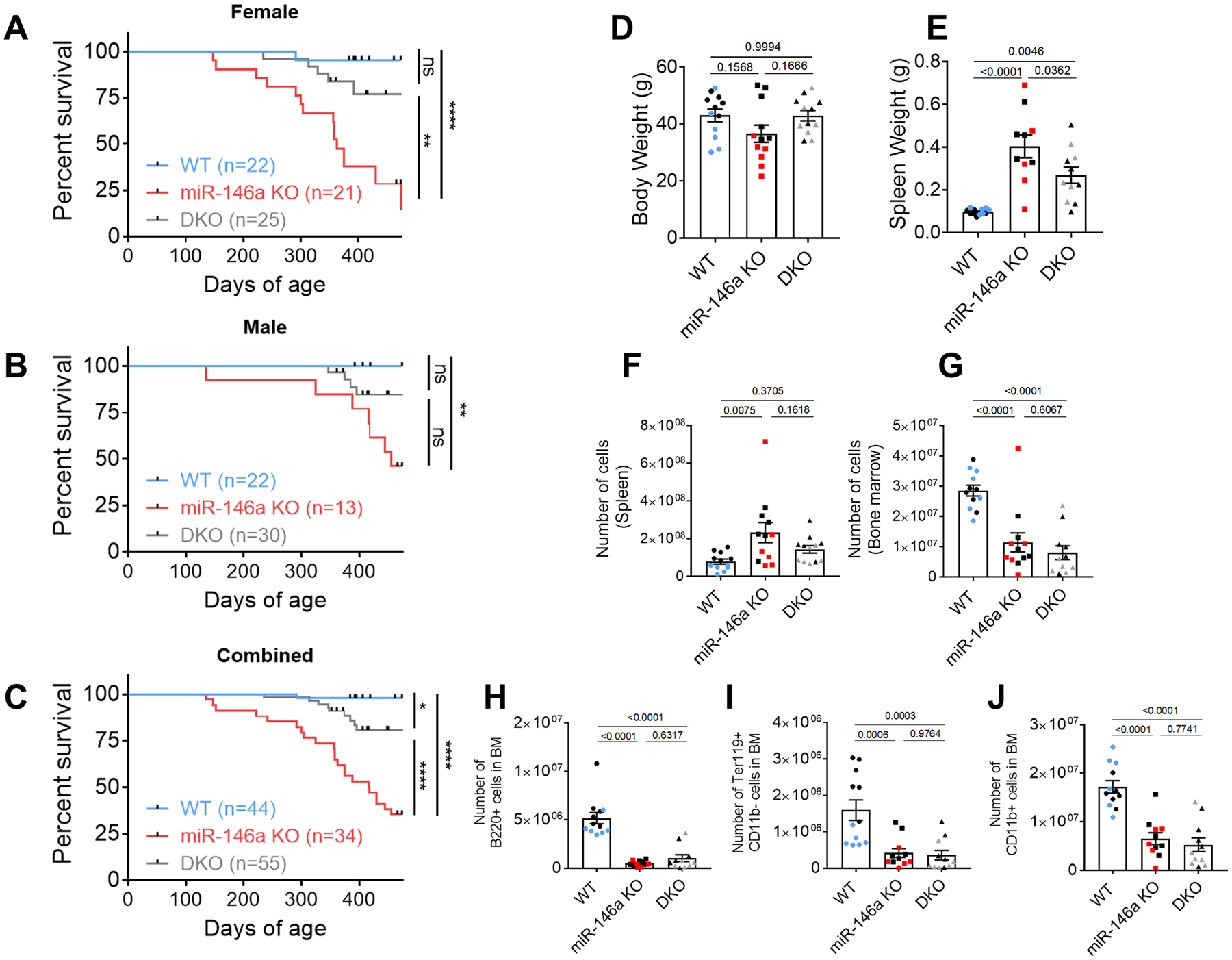
Decreased survival observed in A. miR-146a−/− (KO) female, B. male, C. combined groups of mice is partially rescued by the T cell-specific deletion of miR-155 (miR-146a KO/miR-155 TCKO double knockout, DKO). Log-rank test was used in comparisons, and the p values are summarized (ns, not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001). D. Loss of miR-146a during aging is associated with lower body weight which is rescued back to wild type (WT) levels in DKO group. E. Splenomegaly in aged miR-146a−/− mice is reduced in DKO mice. F. Increased spleen cellularity was also rescued in DKO mice. G. T cell-specific miR-155 does not impact bone marrow (BM) failure in aged miR-146a. H-J. Aged miR-146a−/− BM contains reduced numbers of H. B220+ B cells, I. Ter119+ erythroid progenitors, and J. CD11b+ myelocytes which were not rescued by the deletion of T cell miR-155. In panels D-J, combined data from ~15-months-old male (black) and female mice are shown. Individual measurements and the mean+/− standard error per biological group are shown. P-values from ANOVA were corrected by the Tukey method.
miR-146a and miR-155 control the systemic immune landscape during aging and regulate T cell activation states
To study how miR-146a and miR-155 control the immune response in the context of aging, we characterized the transcriptome of individual immune cells in the spleen using single cell RNA sequencing (scRNAseq). CD45+ splenocytes were sorted via flow cytometry from young WT (n=3) and aged (~15 months) WT (n=6), miR-146a−/− (n=6), and DKO (n=6) male mice, pooled per each group, and subjected to scRNAseq via 10X Genomics technology. Aggregate analysis of 4 groups revealed 16 unique single cell clusters across lymphoid and myeloid lineages that exhibited dynamic changes with aging and between genotypes (Figure 2a). These single cell clusters were annotated using differential expression analyses and with the help of an algorithm we developed which compares gene expression signatures of single cell clusters with the gene expression data from isolated immune cells found in the Immunological Genome Project database (ImmGen, www.immgen.org) (33). Supporting previous findings (1), the frequency of naïve T cell subsets were found to decrease with aging in WT mice while both CD4+ and CD8+ memory T cell subsets were elevated (Figure 2a, ,b).b). The loss of miR-146a in aged mice was associated with further dynamic changes, and T cell-specific deletion of miR-155 in miR-146a−/− animals blunted some of these alterations suggesting miR-146a and miR-155 have opposing functions within the immune system during aging (Figure 2b). The miR-146a and miR-155-dependent changes were observed in multiple cell subsets although some of the cell populations did not differ appreciably (Figure 2b, ,c).c). These findings suggest that miR-146a functions to dampen immune cell activation as a consequence of aging, and that T cell miR-155 is a key mediator of many, but not all, aspects of inflammation in the absence of miR-146a.
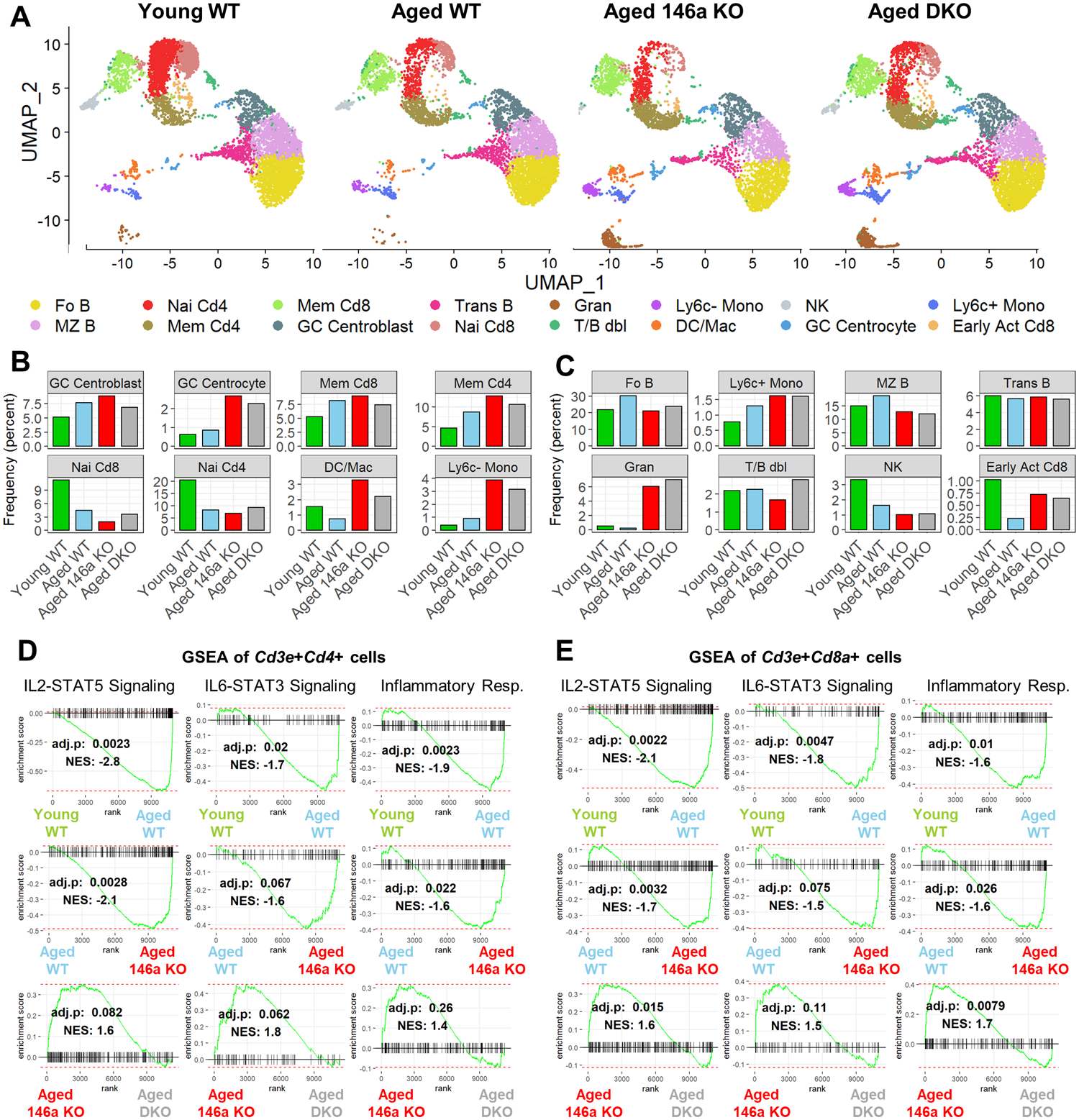
A. Uniform manifold approximation and projection (UMAP) plots showing single cell clusters within the spleens of 15-months-old male mice. Aging is associated with an increase in memory, and a decrease in naïve T cell populations. Loss of miR-146a further amplifies these changes which were mostly reversed in DKO group. B. Cell populations undergoing age- and miR-146a-dependent changes are shown. C. Cell populations that do not show a striking difference between experimental groups are shown. D. Gene set enrichment analysis (GSEA) in Cd3e+Cd4+ T cells showing enrichment of inflammatory pathways in aged miR-146a−/−mice. Inflammatory gene signatures were reversed by T cell-specific deletion of miR-155. E. GSEA of Cd3e+Cd8a+ T cells showing increased inflammation during aging and in the absence of miR-146a. DKO group has lower inflammation compared to miR-146a−/− counterparts. The Benjamini-Hochberg corrected p-values and normalized enrichment score (NES) are shown. GC, germinal center; Mem, memory; Nai, naïve; DC, dendritic cell; Mac, macrophage; Mono, monocyte; Fo B, follicular B; MZ B, marginal zone B; Trans B, transitional B; Gran, granulocyte; T/B dbl, T/B doublet; NK, natural killer; Early Act Cd8, early activated Cd8+ T cell.
We next performed gene set enrichment analysis (GSEA) to investigate the transcriptional activity in immune-related pathways at the single cell level. For this analysis, we utilized all the cells in our dataset expressing Cd3e and Cd4 or Cd3a and Cd8a simultaneously to focus on CD4+ and CD8+ T cells respectively. The expression of IL2-STAT5 signaling pathway genes was significantly enriched upon aging in WT mice in both CD4+ and CD8+ T cell subsets (Figure 2d, ,e).e). Genetic deletion of miR-146a resulted in a further enrichment of this pathway compared to aged WT samples, while T cell-specific loss of miR-155 significantly blunted this enrichment in T cells (Figure 2d, ,e).e). We observed the same patterns when GSEA was performed using IL6-STAT3 signaling and inflammatory response gene sets (obtained from Molecular Signatures Database) (40), suggesting that miR-146a suppresses cytokine signaling and inflammation in aging T cells while miR-155 exacerbates the inflammatory processes during aging.
The lack of miR-146a results in the expansion of Tfh cells in a miR-155-dependent manner
Next, to characterize the immune phenotype in these mice and validate our findings from scRNAseq, we analyzed splenocytes using flow cytometry. As observed in middle-aged (~7 months-old) mice, we found increased overall numbers of CD4+ T cells (Figure 3a), activated CD4+ T cells (Figure 3b, ,c,c, S1a), and reduced numbers of naïve CD4+ T cells (Figure 3d) in miR-146a−/− spleens compared to WT and DKO counterparts at old age (~15 months-old). In addition, we observed increased numbers of Tfh cells in miR-146a−/− spleens which were rescued by T cell-specific deletion of miR-155 (Figure 3e, ,f,f, S1b). Interestingly, we also observed a trending increase in the overall numbers of CD8+ T cells upon deletion of miR-146a (Figure 3g), and a significant increase in the activated subset of these cells which was rescued back to the WT levels (Figure 3h) in the double knockout group. Similarly, the reduced total numbers of naïve CD8+ T cells in miR-146a−/− mice were fully rescued by the T cell-specific deletion of miR-155 (Figure 3i). The number of GC B cells were also found to increase in miR-146a−/− spleens in a T cell miR-155-dependent manner (Figure 3j, S1c). These trends were also evident when the frequencies were assessed instead of total numbers (Figure S2), and are consistent with our scRNAseq findings. As aging was shown to be associated with increased levels of autoreactive antibodies (autoantibodies) (20, 41, 42), we next quantified the anti-double stranded DNA (dsDNA) antibodies in the serum of these mice. Aged miR-146a−/− serum had significantly higher levels of IgG specific for anti-dsDNA compared to WT counterparts, and this increase was reversed in DKO mice (Figure 3k). Data from our lab and others have shown loss of miR-146a or constitutive expression of miR-155 results in a myeloproliferative phenotype (23, 30, 39). When we examined the spleens of aged mice, we noted significantly higher numbers of CD11b+ myeloid cells in miR-146a−/− animals which was reduced back to WT levels in DKO counterparts (Figure 3l). These differences were comparable to the observations in middle-aged mice (20), suggesting elevated Tfh cells, GC B cells, myeloproliferation, and autoantibodies are maintained in elderly mice. Altogether, these data suggest that deleting miR-155 in T cells prevents the expansion of activated Tfh cells and autoimmunity in aged, chronically inflamed miR-146a−/− mice.
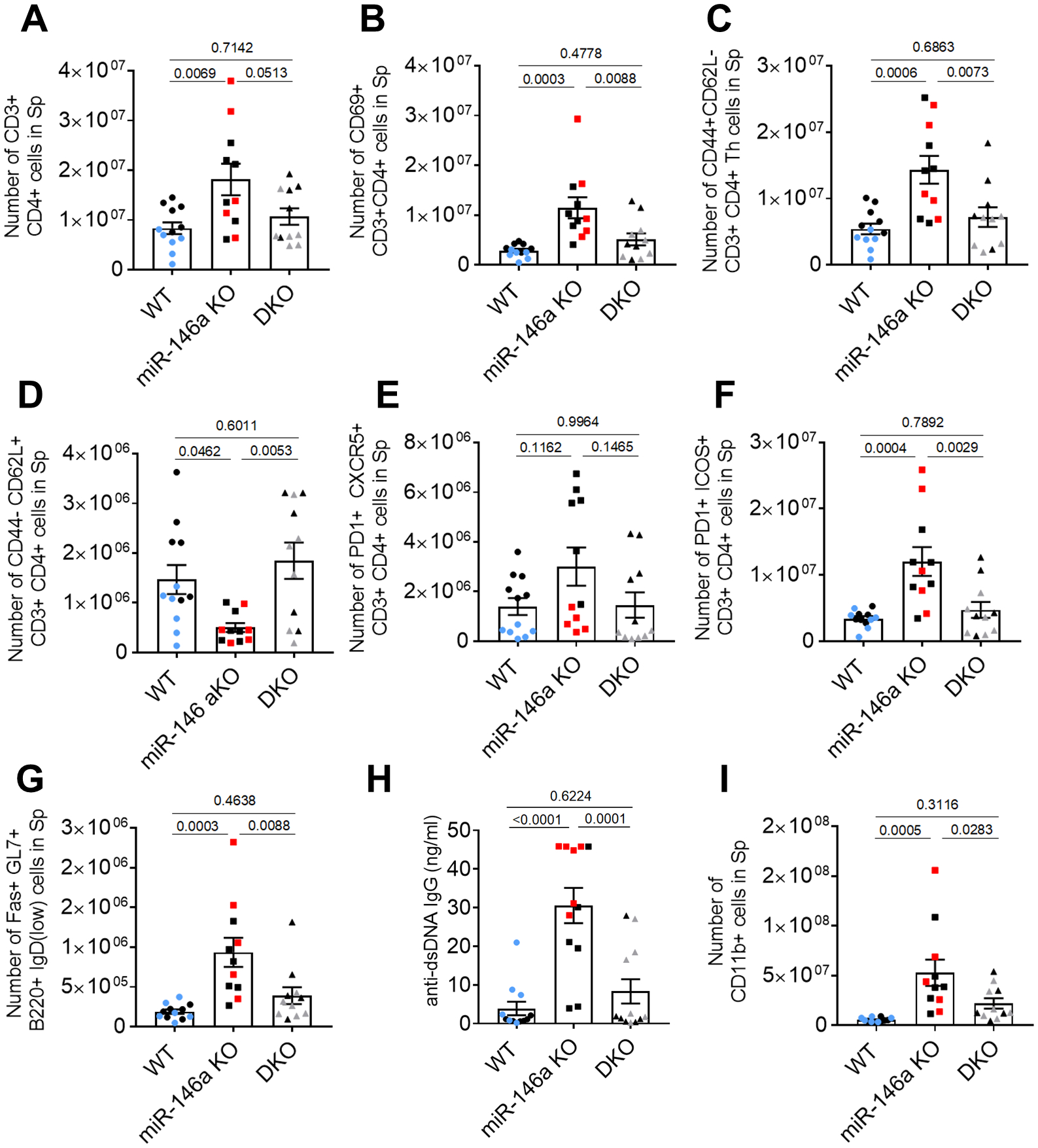
Flow cytometric analysis of spleen showing total numbers of A. CD3+CD4+ T helper (Th) cells, B. CD69+ activated Th cells, C. CD44+CD62L- activated Th cells, and D. CD44-CD62L+ naïve Th cells. E. PD1 and CXCR5-expressing T follicular helper (Tfh) cells are elevated in aged miR-146−/− mice which is reduced in DKO group. F. PD1+ICOS+ Tfh cells are also elevated in aged miR-146a mice and are rescued by T cell specific deletion of miR-155. G. The total numbers of CD8+ cells are higher in aged miR-146a−/− mice. H. The number of the activated CD8+ T cells increases upon loss of miR-146a in a miR-155 dependent manner. I. Lower numbers of naïve CD8+ T cells in miR-146a−/− mice were reversed back to WT levels in DKO group. J. Germinal center B cells are elevated in miR-146a−/− spleens suggesting an increased T-B cell crosstalk. K. ELISA results showing increased levels of double stranded DNA (dsDNA) reacting autoantibodies in aged miR-146a KO mice. Autoantibody levels are reduced upon miR-155 deletion in T cells suggesting increased autoantibody production in the absence of miR-146a is mediated by T cell-specific miR-155. L. miR-146a KO mice exhibit myeloproliferation assessed by the increased numbers of CD11b+ splenocytes which is partially rescued upon deletion of T cell miR-155. Data from individual mice are depicted in addition to mean (+/− standard error). Mice in these experiments are 15-months old and black points indicate males. Tukey-corrected p-values from ANOVA tests are shown between groups.
Loss of miR-146a promotes IgG antibody responses against antigens from multiple tissue types and is largely reversed by T cell-specific deletion of miR-155
Autoantibodies can be generated against various self-tissue antigens, especially in the context of aging (43). To identify if autoantibodies are raised against tissue antigens in ~15 months-old miR-146a−/− mice, we performed western blots on various tissues from WT, miR-146a−/−, DKO and Rag−/− mice. In these assays, the whole organ protein lysates were size fractionated using denaturing polyacrylamide gel electrophoresis (SDS-PAGE), transferred to membranes, and subsequently probed with the serum collected from different groups of aged mice as the primary antibody, followed by anti-mouse IgG secondary antibodies to reveal tissue proteins bound by serum autoantibodies. In these experiments, organs and serum collected from Rag−/− mice serve as negative controls since these mice cannot produce antibodies due to the lack of B and T cells (44), and β-actin ensures similar amounts of protein loading in these assays. Consistent with the anti-dsDNA autoantibody data, miR-146a−/− mouse serum exhibited increased reactivity to multiple tissue antigens in brains (Figure 4a), kidneys (Figure 4b), and spleens (Figure 4c) at old age. Deletion of miR-155 in T cells reduced auto-reactivity back to levels comparable to aged WT mice in most tissues. Interestingly, we did not observe a global decrease in autoantibodies against liver self-antigens in DKO serum (Figure 4d), although a few targets were still diminished upon deletion of miR-155 in T cells, indicating that lack of miR-155 in T cells can block autoantibody production against certain liver tissue antigens. Importantly, the increase in autoantibodies in miR-146a−/− mice was not due to increases in total antibody production (Figure S3). Altogether, these findings suggest that loss of miR-146a during aging results in elevated levels of autoantibodies against self-tissue antigens across multiple organs, and the increased autoantibody production can be suppressed by deleting miR-155 specifically in T cells.
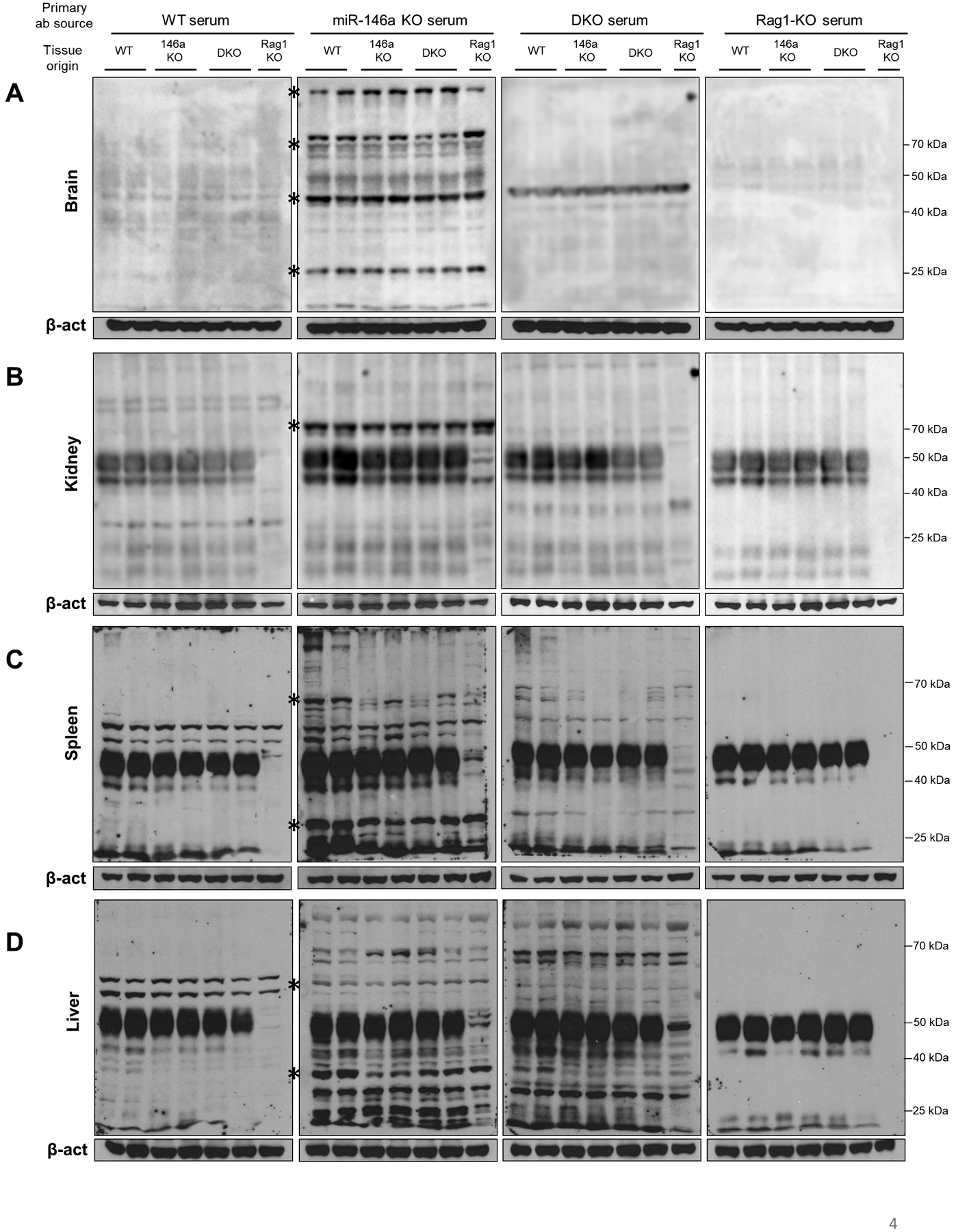
Western blotting was performed on whole organ extracts using serum from various groups of aged mice (~15 months-old) as primary antibody source. Autoreactive antibodies were then detected using anti-mouse IgG antibody. Rag1-KO tissue in the blots serve as a control in which no background antibody is present due to lack of adaptive immunity. Panels labeled as Rag1-KO serum serve as a negative control for primary antibody; therefore, the positive signal in these blots is attributable to antibodies already present in the tissue extract (i.e. tissue-bound antibodies, or IgG on B cells within the organ). Similar levels of beta-actin (β-act) ensures consistent protein loading in the assay. A. Loss of miR-146a is associated with increased autoantibodies reacting to tissue antigens in brain. The majority of autoreactivity was lost upon T cell specific deletion of miR-155, as denoted by the asterisks. Increased serum reactivity to self-antigens was also observed in B. kidney, C. spleen, and D. liver, which was reduced in DKO serum, suggesting a multi-organ autoimmunity in aging miR-146a−/− animals is mediated by the T cell-specific miR-155.
miR-146a−/− T cells have enhanced aerobic glycolysis which is reversed upon loss of miR-155
T cell activation is characterized by an increased metabolic output and a highly glycolytic state (45, 46). To investigate the metabolic changes in T cells in the process of aging, we first analyzed our previously published RNAseq data obtained from splenic CD4+ T cells sorted from middle-aged (~7 months-old) WT and whole-body miR-146a−/−, miR-155−/−, and double knockout (DKO) mice (accession: {"type":"entrez-geo","attrs":{"text":"GSE58373","term_id":"58373"}}GSE58373) (20). In our previous study, CD4+ T cells from miR-146a−/− mice were found to have an enhanced activation phenotype with more profound Tfh differentiation (20). When the metabolic pathways were examined in these cells, we observed a significant enrichment of the glycolysis gene set in aged miR-146a−/− CD4+ T cells compared to aged WT counterparts (Figure 5a). Glycolysis gene signature was also enriched in miR-146a−/− T cells when compared to DKO group suggesting that the increased glycolysis in miR-146a−/− T cells is mediated by miR-155. Supporting this notion, when CD4+ T cells from aged WT and aged DKO were compared to each other, the glycolysis signature was not differentially enriched, indicating the deletion of miR-155 on miR-146a−/− background reverses the increased glycolysis back to WT levels. Consistent with these findings, the expression levels of genes regulating the glycolysis pathway including AldoA, Hif1a, Hk2 and Slc2a6 (Glut6) were higher in miR-146a−/− T cells and were rescued by DKO (Figure 5b). These data suggest that T cells in middle-aged miR-146a−/− mice have increased glycolytic activity that is promoted by miR-155, and these miRNAs have opposing functions in regulating T cell activation and metabolism in the context of aging.
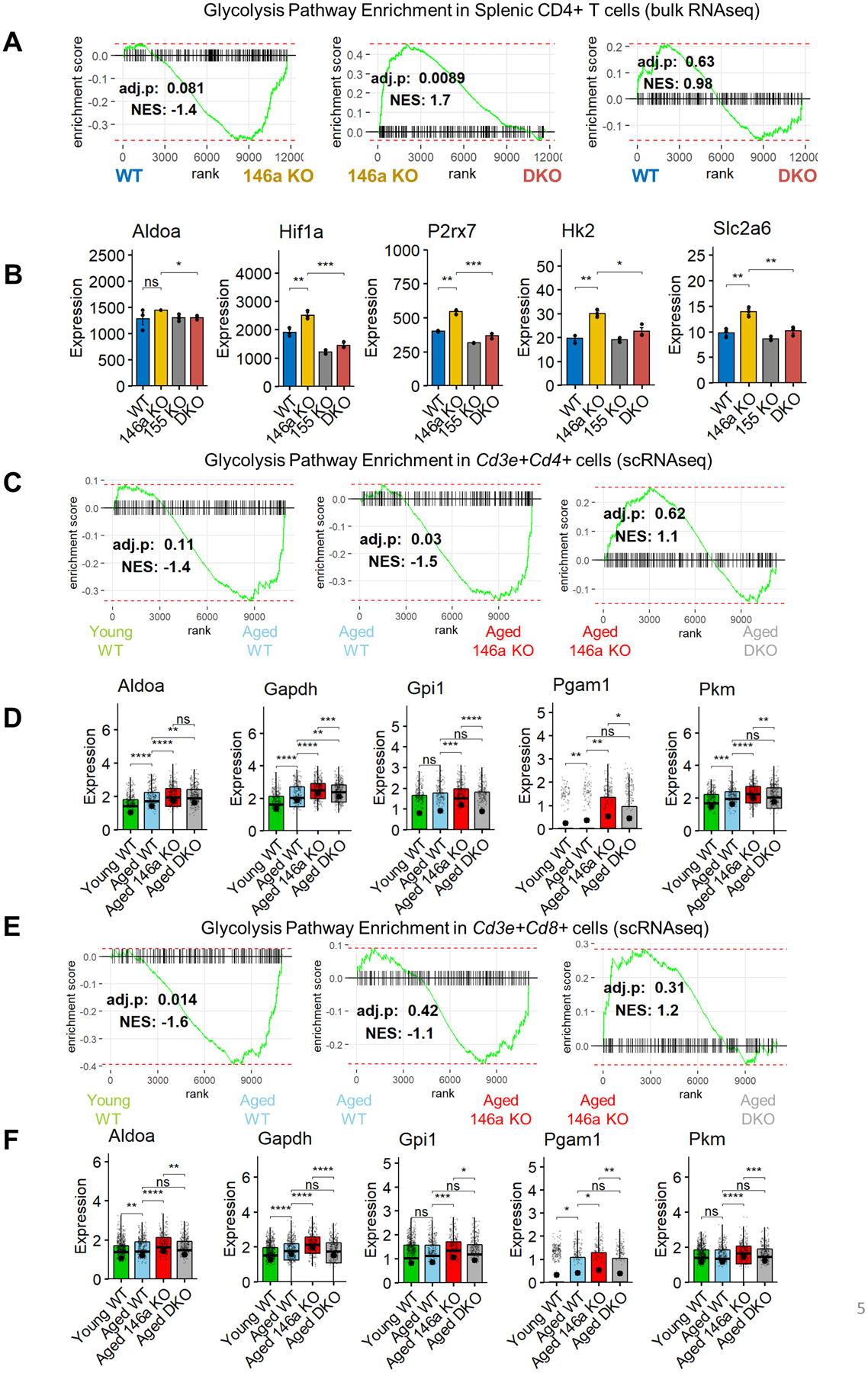
A. RNA sequencing of splenic CD4+ T cells from middle aged (~7 months) mice (20) reveals an increased glycolysis gene signature in miR-146−/− cells. This enrichment was mostly rescued in DKO mice. B. Glycolytic gene expression was reduced when miR-155 was knocked out globally. Holm-corrected t tests were used in comparisons (n=3 per group). C. Glycolysis pathway enrichment in miR-146a−/− mice was sustained in old age (~14–15 months) (scRNAseq data, Figure 2), which was rescued upon T cell-specific deletion of miR-155. D. Boxplots showing the glycolytic gene expression within Cd3e+Cd4+ cells. Data from individual cells are shown along with average expression per group (large black point). E. Glycolysis gene signature is enriched in Cd3e+Cd8a+ T cells with aging and further in miR-146a−/− mice. Deletion of miR-155 in T cells reversed this effect. F. The differential expression of the glycolysis pathway genes was blunted in DKO group. The Holm-corrected p values of Wilcoxon tests are represented by asterisks (ns, not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001).
We then assessed whether the increased glycolysis phenotype is maintained in older mice (~15 months-old) using our scRNAseq dataset (Figure 2). For these analyses, we divided our dataset into CD3e+CD4+ and CD3e+CD8a+ cells, as done previously (Figure 2d, ,e),e), and performed GSEA using the glycolysis gene set. Supporting our previous findings from sorted T cells, scRNAseq indicated increased levels of CD4+ T cell glycolysis with aging, a phenotype which was exacerbated on a miR-146a−/− background (Figure 5c). CD4+ T cells from DKO mice showed reduced expression of glycolysis pathway genes compared to miR-146a−/− counterparts although this trend was not statistically significant. Expression levels of several genes in glycolysis pathway showed an age-dependent increase in WT cells that was further enhanced in miR-146a−/− mice and subsequently reversed by DKO (Figure 5d). Age and miR-146−/− dependent increase in glycolysis signature within CD4+ T cells was also observed in CD8+ T cells (Figure 5e). Multiple genes in the glycolysis pathway were upregulated in aged miR-146a−/− CD8+ T cells and were subsequently rescued by concomitant deletion of miR-155 (Figure 5f). These data suggest that T cell-specific miR-155 is important for maintaining elevated glycolysis in aged miR-146a−/− T cells, and the balance between miR-146a and miR-155 regulates T cell metabolism and inflammation during aging.
Deletion of Mpc1 in T cells results in the enrichment of activated T and Tfh cells during aging
Since our data suggest that miR-146a-deficient T cells have enhanced aerobic glycolysis which can be reversed by deleting miR-155, we next wanted to determine if skewing T cells towards aerobic glycolysis was sufficient to promote the expansion of Tfh and GC B cells in the context of aging. To address this point, we utilized a newly developed transgenic mouse strain in which floxed mitochondrial pyruvate carrier 1 (Mpc1 fl/fl) is deleted only in T cells by the CD4-Cre lineage driver. Mpc1 is critical for the mitochondrial uptake of pyruvate generated in the cytoplasm as the end product of glycolysis (47, 48). Because of the inability to transfer pyruvate into the mitochondria, Mpc1 knock-out animals exhibit increased aerobic glycolysis to meet the cellular energetic demand (47, 48). To study the role of aerobic glycolysis in T cell aging, we immunophenotyped splenocytes collected from T cell specific Mpc1−/− mice and floxed controls that were aged for ~15 months. Skewing T cell metabolism toward aerobic glycolysis by deleting Mpc1 increased the levels of activated CD4+ T cells and reduced the levels of naïve T cells (Figure 6a). A similar increase in the activated subset and a decrease in the naïve subset were also evident in CD8+ T cell compartment (Figure 6b). When we examined Tfh markers including PD1, CXCR5, and ICOS, we observed a statistically significant increase upon T cell-specific loss of Mpc1 (Figure 6c, S4a). Consistent with the changes in Tfh frequency, we also observed increased proportions of GC B cells (Figure 6d, S4b). Importantly, unlike the aged mice, the frequency of Tfh cells were similar in young (~8 weeks old) Mpc1−/− mice (Figure 6e). Altogether, these data suggest that skewing T cells towards aerobic glycolysis can lead to increased Tfh and GC B cell expansion, similar to miR-146a−/− mice, in the context of aging.
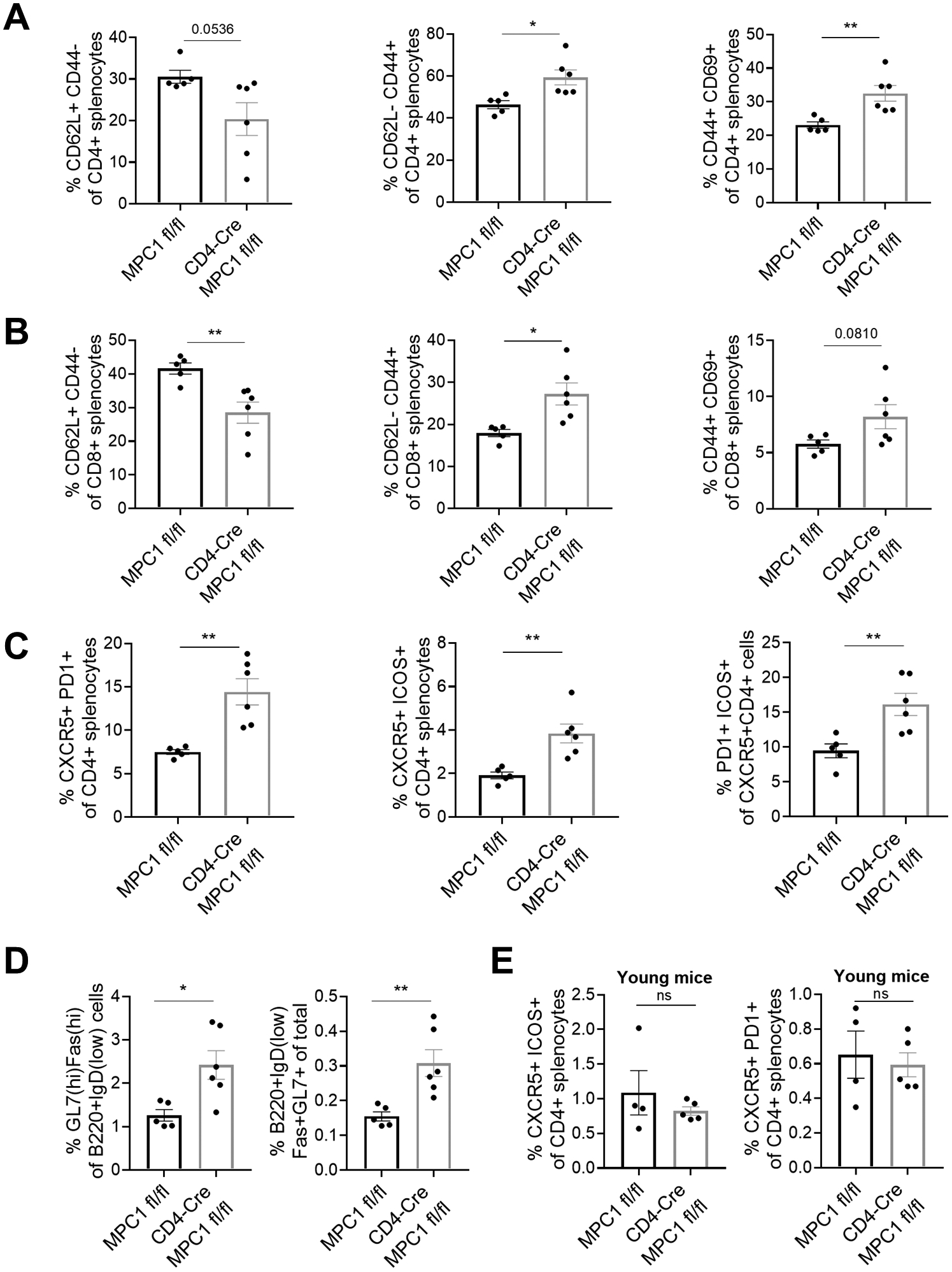
A. Deletion of Mpc1 in T cells is associated with reduced levels of naïve and increased levels of activated CD3+CD4+ splenic T cells during aging (~15 month-old) as assessed by flow cytometry. B. Splenic CD8+ T cells exhibit a similar activation phenotype in aged Mpc1 knockout mice. C. Deletion of Mpc1 results in an increased frequency of Tfh cells during aging. D. Splenic GC B cells are elevated in aged Mpc1 knockout mice. E. In young mice (8–12 weeks old), the deletion of MPC1 did not increase the frequency of Tfh cells, suggesting the effect of enhanced glycolysis on Tfh phenotypes is dependent on the aging process. The p-values of student’s t-test tests are represented by asterisks (ns, not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001).
Discussion
In the current study, we identified a novel interaction between miR-146a and miR-155 in healthy aging and longevity. Our findings, like others, demonstrate that miR-146a is critical for a normal murine lifespan, as the genetic deletion of miR-146a results in a multi-organ autoimmune response with the involvement of Tfh cells and accelerated death. Importantly, deleting miR-155 in T cells is sufficient to reverse many of the negative effects observed in miR-146a−/− animals, including increased mortality. However, the loss of T cell-specific miR-155 did not completely rescue the lifespan of miR-146a−/− mice, which could be at least in part attributed to the inability to restore bone marrow cellularity and composition. Additionally, deleting miR-146a globally is expected to have non-T cell effects that likely also contribute to shortened lifespan. Indeed, multiple studies have shown that miR-146a functions in a variety of cell types to regulate immune responses (21, 23, 37–39, 49). Thus, while T cell-expressed miR-155 strongly contributes to inflammatory disease during aging in this model, it is not sufficient to mediate all aspects of inflammatory disease observed in miR-146a−/− mice. An interesting connection between miR-146a and miR-155 was found by studies suggesting that miR-155 expression is elevated in miR-146a-deficient T cells and can contribute to overactivation of the immune system (21, 37). Altogether, our observations in this study provide strong evidence that miR-155 in T cells can have an adverse effect on health and survival during aging, at least in settings of chronic inflammation. Further, maintaining the proper balance between miR-146a and miR-155 is critical for effective immunity against infection and cancer, where miR-155 is protective, and in the context of age-related autoimmunity, where miR-155 promotes pathology and impacts lifespan.
The essential roles of miRNAs in regulating T cell differentiation and function have been clearly established by prior work (50–57). Our study reveals an interesting link between the miRNA regulatory networks and cellular metabolism in shaping T cell responses in the context of aging and autoimmunity. Cellular metabolism is increasingly appreciated to be not only critical for mediating immune effector function, but also for determining immune cell fate and activation states (58–61). Previously, we identified increased glycolysis in macrophages deficient in miR-146a due to increased TRAF6 and mTOR signaling (37). Here, we provide evidence that miR-146a-deficient T cells in aged mice also have enhanced aerobic glycolysis compared to WT counterparts, and that the removal of miR-155 can reverse this phenotype, suggesting that the metabolic state of T cells is a critical determinant in age-mediated inflammation. Our data are consistent with studies in cancer and non-hematopoietic cells, where miR-155 was found to promote glycolysis through mechanisms involving targeting of C/EBPβ, HDAC4, SOCS1 and PIK3R1 (62–64). Future work will explore whether miR-155-mediated promotion of glycolysis in T cells during aging occurs through these direct targets or via a unique mechanism that has not yet been described.
Several studies have recently shown that cellular metabolism influences T cell differentiation and effector responses (59, 60, 65–68). In the context of Tfh biology, metabolism plays a complex role in regulating the cellular phenotype. Some studies suggested that the Tfh master transcription factor Bcl6 represses the expression of hexokinase 2 (Hk2) and other key metabolic genes, and that Tfh cells exhibit lower glycolytic activity compared to Th1 cells during infection (69, 70). On the contrary, others reported higher levels of glycolysis in Tfh cells compared to non-Tfh counterparts, and overexpressing the glucose transporter Slc2a1 on T cells was sufficient to increase steady-state Tfh and GC B cell numbers (71, 72). This discrepancy between different studies may be explained in part by the observation that autoreactive Tfh cells were highly sensitive to glycolysis inhibition while virus-induced Tfh cells remained unimpacted, indicating that the inflammatory context matters in Tfh biology and increased glycolysis is critical for the development of autoreactive Tfh cells (72). Our observations are in accordance with these latter findings and suggest that miR-146a and miR-155 controls the development of autoreactive Tfh cells through regulating glycolysis. While it is widely accepted that activated T cells require higher glycolytic activity to support proliferation and effector cytokine production (66, 73, 74), our data demonstrate that skewing T cell metabolism toward aerobic glycolysis by deleting Mpc1 is sufficient to promote their activation in the context of age-mediated inflammation. This is also consistent with a recent study which showed that fissed punctate mitochondria favoring aerobic glycolysis lead to the development of effector T cells in bacterial infection models (75). Our study suggests that increased aerobic glycolysis in T cells could potentially increase morbidity in the elderly, and future studies will continue to investigate Mpc1 and metabolism-altering miRNAs in the context of aging and longevity.
In conclusion, our data highlight the importance of maintaining proper miRNA regulation in immune cells to promote healthy aging and long-term survival. Specifically, interventions targeting miR-155 expression and function in T cells may have the potential to prevent co-morbidities of aging, extend lifespan, and improve the quality of life in the elderly, at least in some circumstances. Further pre-clinical and clinical studies will be essential to realize the potential of manipulating miRNAs in the immune system to tackle age-related disease.
Key Points
- miR-155 in T cells contributes to the shortened lifespan of miR-146a−/− mice.
- Autoimmunity in aged miR-146a−/− mice is mediated in part by T cell miR-155.
- Aerobic glycolysis is associated with increased Tfh and GC B cells during aging.
Acknowledgements
We thank the University of Utah High Throughput Genomics (Opal Allen, Brian Dalley) and Bioinformatics (Chris Stubben, Chris Conley) core facilities for the help with RNAseq and scRNAseq data analysis. We would also like to thank the University of Utah Flow Cytometry core facility (James Marvin) for assistance with flow cytometric analysis and cell sorting.
Funding: This work was supported by the National Institutes of Health (NIH) grants R01AG047956 and R01AI123106 (RMO), R01CA166450 and R21AI132869 (DSR), and Training Grants T32 AI055434 (AGR) and T32 AI138945-1 (KMB).
Footnotes
Competing Interests: Authors declare no competing interests exist.
Data availability: High throughput scRNAseq data generated in this manuscript is available at Gene Expression Omnibus ({"type":"entrez-geo","attrs":{"text":"GSE138222","term_id":"138222"}}GSE138222). URL: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc={"type":"entrez-geo","attrs":{"text":"GSE138222","term_id":"138222"}}GSE138222